Back
BackProblem 1a
Why did geneticists believe, even before direct experimental evidence was obtained, that the genetic code would turn out to be composed of triplet sequences and be nonoverlapping? Experimentally, how were these suppositions shown to be correct?
Problem 1b
What experimental evidence provided the initial insights into the compositions of codons encoding specific amino acids?
Problem 1c
How were the specific sequences of triplet codes determined experimentally?
Problem 1d
How were the experimentally derived triplet codon assignments verified in studies using bacteriophage MS2?
Problem 2
Write a short essay that summarizes the key properties of the genetic code and the process by which RNA is transcribed on a DNA template.
Problem 3
Assuming the genetic code is a triplet, what effect would the addition or loss of two nucleotides have on the reading frame? The addition or loss of three, six, or nine nucleotides?
Problem 4
The mRNA formed from the repeating tetranucleotide UUAC incorporates only three amino acids, but the use of UAUC incorporates four amino acids. Why?
Problem 5
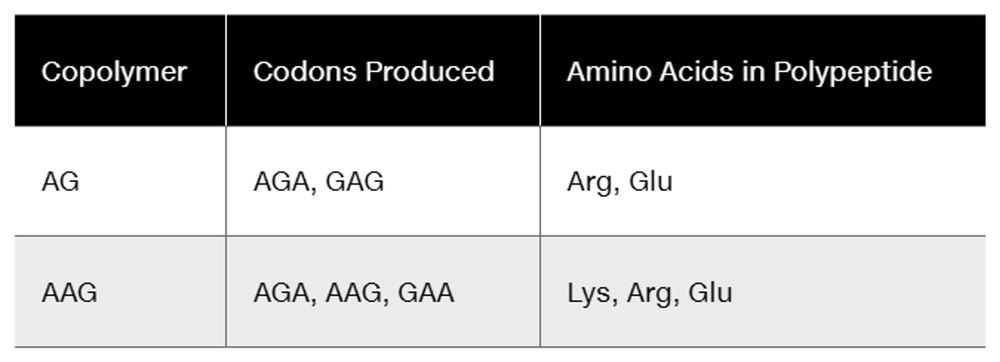
In studies using repeating copolymers, AC . . . incorporates threonine and histidine, and CAACAA . . . incorporates glutamine, asparagine, and threonine. What triplet code can definitely be assigned to threonine?
Problem 6
In a coding experiment using repeating copolymers, the following data were obtained:
AGG is known to code for arginine. Taking into account the wobble hypothesis, assign each of the four codons produced in the experiment to its correct amino acid.
Problem 7
In the triplet binding technique, radioactivity remains on the filter when the amino acid corresponding to the codon is labeled. Explain the rationale for this technique.
Problem 8
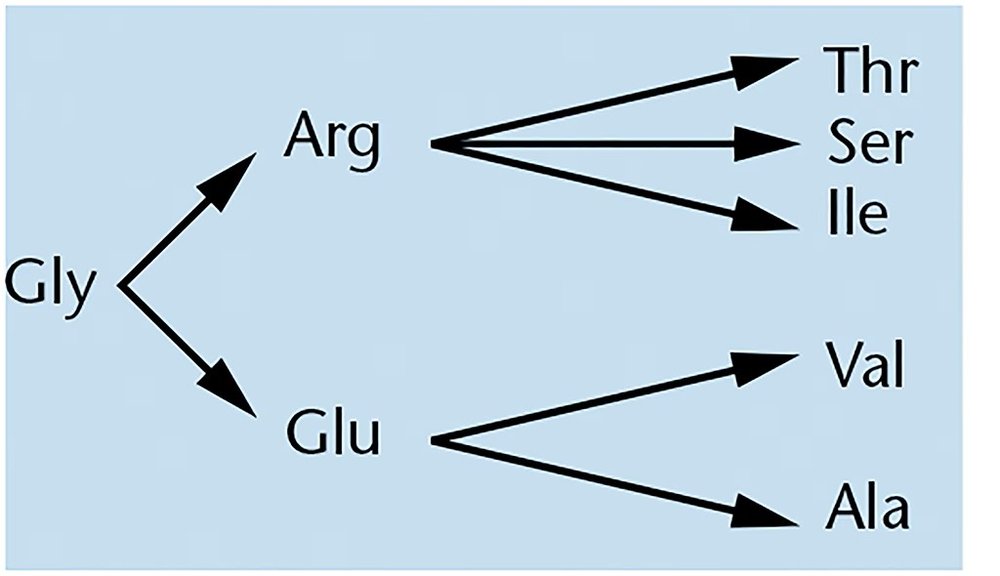
When the amino acid sequences of insulin isolated from different organisms were determined, differences were noted. For example, alanine was substituted for threonine, serine for glycine, and valine for isoleucine at corresponding positions in the protein. List the single-base changes that could occur in codons of the genetic code to produce these amino acid changes.
Problem 9
In studies of the amino acid sequence of wild-type and mutant forms of tryptophan synthetase in E. coli, the following changes have been observed:
Determine a set of triplet codes in which only a single-nucleotide change produces each amino acid change.
Problem 10
Why doesn't polynucleotide phosphorylase (Ochoa's enzyme) synthesize RNA in vivo?
Problem 11
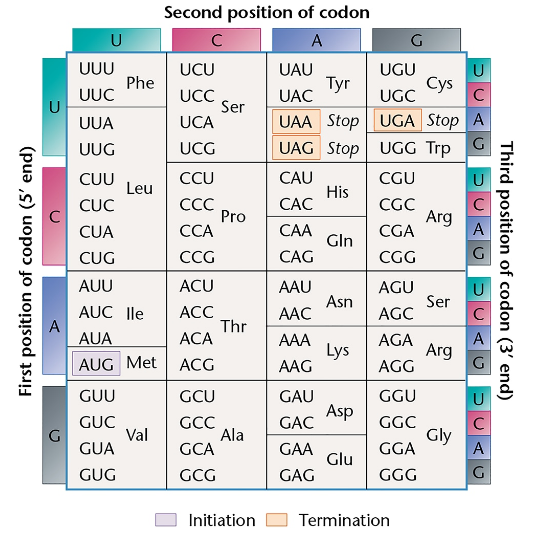
Refer to Table 13.1. Can you hypothesize why a synthetic RNA composed of a mixture of poly U poly A would not stimulate incorporation of ¹⁴C-phenylalanine into protein?
Problem 12
Predict the amino acid sequence produced during translation by the following short hypothetical mRNA sequences (note that the second sequence was formed from the first by a deletion of only one nucleotide):
Sequence 1: 5'-AUGCCGGAUUAUAGUUGA-3'
Sequence 2: 5'-AUGCCGGAUUAAGUUGA-3'
What type of mutation gave rise to sequence 2?
Problem 13a
A short RNA molecule was isolated that demonstrated a hyperchromic shift, indicating secondary structure. Its sequence was determined to be
5'-AGGCGCCGACUCUACU-3'
Propose a two-dimensional model for this molecule.
Problem 13b
A short RNA molecule was isolated that demonstrated a hyperchromic shift, indicating secondary structure. Its sequence was determined to be
5'-AGGCGCCGACUCUACU-3'
What DNA sequence would give rise to this RNA molecule through transcription?
Problem 13c
A short RNA molecule was isolated that demonstrated a hyperchromic shift, indicating secondary structure. Its sequence was determined to be
5'-AGGCGCCGACUCUACU-3'
If the molecule were a tRNA fragment containing a CGA anticodon, what would the corresponding codon be?
Problem 13d
A short RNA molecule was isolated that demonstrated a hyperchromic shift, indicating secondary structure. Its sequence was determined to be
5'-AGGCGCCGACUCUACU-3'
If the molecule were an internal part of a message, what amino acid sequence would result from it following translation?
Problem 14
A glycine residue is in position 210 of the tryptophan synthetase enzyme of wild-type E. coli. If the codon specifying glycine is GGA, how many single-base substitutions will result in an amino acid substitution at position 210? What are they? How many will result if the wild-type codon is GGU?
Problem 15a
Shown here is a hypothetical viral mRNA sequence:
5'-AUGCAUACCUAUGAGACCCUUGGA-3'
Assuming that it could arise from overlapping genes, how many different polypeptide sequences can be produced? What are the sequences?
Problem 15b
Refer to the genetic coding dictionary to respond to the following:
A base-substitution mutation that altered the sequence shown in part (a) eliminated the synthesis of all but one polypeptide. The altered sequence is shown here:
5'-AUGCAUACCUAUGUGACCCUUGGA-3'
Determine why.
Problem 16
Most proteins have more leucine than histidine residues, but more histidine than tryptophan residues. Correlate the number of codons for these three amino acids with this information.
Problem 17
Define the process of transcription. Where does this process fit into the central dogma of molecular biology (DNA makes RNA makes protein)?
Problem 18
What observations suggested the existence of mRNA?
Problem 19
Describe the structure of RNA polymerase in bacteria. What is the core enzyme? What is the role of the σ subunit?
Problem 20
Write a paragraph describing the abbreviated chemical reactions that summarize RNA polymerase-directed transcription.
Problem 21
Messenger RNA molecules are very difficult to isolate in bacteria because they are rather quickly degraded in the cell. Can you suggest a reason why this occurs? Eukaryotic mRNAs are more stable and exist longer in the cell than do bacterial mRNAs. Is this an advantage or a disadvantage for a pancreatic cell making large quantities of insulin?
Problem 22
Present an overview of various forms of posttranscriptional RNA processing in eukaryotes. For each, provide an example.
Problem 23
One form of posttranscriptional modification of most eukaryotic pre-mRNAs is the addition of a poly-A sequence at the 3' end. The absence of a poly-A sequence leads to rapid degradation of the transcript. Poly-A sequences of various lengths are also added to many bacterial RNA transcripts where, instead of promoting stability, they enhance degradation. In both cases, RNA secondary structures, stabilizing proteins, or degrading enzymes interact with poly-A sequences. Considering the activities of RNAs, what might be general functions of 3'-polyadenylation?