Hey, everyone. So in this video, we're going to talk about the beginnings of DNA replication. Now, under DNA replication, a template strand is used to synthesize a new DNA strand that is complementary to it. Now, here, we're going to say that our template strand, well, this is just the starting or parent strand that is copied during replication. So, if we take a look here, our template strand is this right here and this is actually a DNA Double Helix made up of 2 template strands. Where we're going to say our complementary or our daughter strand, well, this is the newly synthesized strand of DNA copied from the template strand. So here we have our DNA double helix made up of 2 template strands, those nitrogenous bases are linked together, remember by hydrogen bonds. What happens is we start to cut through these hydrogen bonds and unwind the DNA, exposing the template strands. So here are 2 step template strands exposed, the nitrogenous bases are no longer hydrogen bonded, and what we start doing is we start copying these exposed template strands. We can see this growing green strand here and this growing green strand here. We're copying the exposed template strands. Over time, we just keep going higher and higher up the DNA double helix, it's becoming more and more unwound, and our daughter strands are growing and growing. Eventually, what we'll get at the end is 2 DNA double helixes. And if you can see, we see that they are a mixture of what? Our template strand and our daughter strand. We're going to say that the 2 new DNAs the double helices or double helices that were created, we're going to conserve some of our old DNA. This is what we call the semi-conservative model, where we say both double helices are going to have 1 template strand and 1 daughter strand. So, in that way, you kind of retain some of the old DNA we had in the very beginning. The starting or parent strand is still present in both double helices as well as our newly copied daughter strands. Alright. So just remember, when we're talking about DNA replication, we're talking about the semi-conservative model.
- 1. Matter and Measurements4h 29m
- What is Chemistry?5m
- The Scientific Method9m
- Classification of Matter16m
- States of Matter8m
- Physical & Chemical Changes19m
- Chemical Properties8m
- Physical Properties5m
- Intensive vs. Extensive Properties13m
- Temperature (Simplified)9m
- Scientific Notation13m
- SI Units (Simplified)5m
- Metric Prefixes24m
- Significant Figures (Simplified)11m
- Significant Figures: Precision in Measurements7m
- Significant Figures: In Calculations19m
- Conversion Factors (Simplified)15m
- Dimensional Analysis22m
- Density12m
- Specific Gravity9m
- Density of Geometric Objects19m
- Density of Non-Geometric Objects9m
- 2. Atoms and the Periodic Table5h 23m
- The Atom (Simplified)9m
- Subatomic Particles (Simplified)12m
- Isotopes17m
- Ions (Simplified)22m
- Atomic Mass (Simplified)17m
- Atomic Mass (Conceptual)12m
- Periodic Table: Element Symbols6m
- Periodic Table: Classifications11m
- Periodic Table: Group Names8m
- Periodic Table: Representative Elements & Transition Metals7m
- Periodic Table: Elemental Forms (Simplified)6m
- Periodic Table: Phases (Simplified)8m
- Law of Definite Proportions9m
- Atomic Theory9m
- Rutherford Gold Foil Experiment9m
- Wavelength and Frequency (Simplified)5m
- Electromagnetic Spectrum (Simplified)11m
- Bohr Model (Simplified)9m
- Emission Spectrum (Simplified)3m
- Electronic Structure4m
- Electronic Structure: Shells5m
- Electronic Structure: Subshells4m
- Electronic Structure: Orbitals11m
- Electronic Structure: Electron Spin3m
- Electronic Structure: Number of Electrons4m
- The Electron Configuration (Simplified)22m
- Electron Arrangements5m
- The Electron Configuration: Condensed4m
- The Electron Configuration: Exceptions (Simplified)12m
- Ions and the Octet Rule9m
- Ions and the Octet Rule (Simplified)8m
- Valence Electrons of Elements (Simplified)5m
- Lewis Dot Symbols (Simplified)7m
- Periodic Trend: Metallic Character4m
- Periodic Trend: Atomic Radius (Simplified)7m
- 3. Ionic Compounds2h 18m
- Periodic Table: Main Group Element Charges12m
- Periodic Table: Transition Metal Charges6m
- Periodic Trend: Ionic Radius (Simplified)5m
- Periodic Trend: Ranking Ionic Radii8m
- Periodic Trend: Ionization Energy (Simplified)9m
- Periodic Trend: Electron Affinity (Simplified)8m
- Ionic Bonding6m
- Naming Monoatomic Cations6m
- Naming Monoatomic Anions5m
- Polyatomic Ions25m
- Naming Ionic Compounds11m
- Writing Formula Units of Ionic Compounds7m
- Naming Ionic Hydrates6m
- Naming Acids18m
- 4. Molecular Compounds2h 18m
- Covalent Bonds6m
- Naming Binary Molecular Compounds6m
- Molecular Models4m
- Bonding Preferences6m
- Lewis Dot Structures: Neutral Compounds (Simplified)8m
- Multiple Bonds4m
- Multiple Bonds (Simplified)6m
- Lewis Dot Structures: Multiple Bonds10m
- Lewis Dot Structures: Ions (Simplified)8m
- Lewis Dot Structures: Exceptions (Simplified)12m
- Resonance Structures (Simplified)5m
- Valence Shell Electron Pair Repulsion Theory (Simplified)4m
- Electron Geometry (Simplified)8m
- Molecular Geometry (Simplified)11m
- Bond Angles (Simplified)11m
- Dipole Moment (Simplified)15m
- Molecular Polarity (Simplified)7m
- 5. Classification & Balancing of Chemical Reactions3h 17m
- Chemical Reaction: Chemical Change5m
- Law of Conservation of Mass5m
- Balancing Chemical Equations (Simplified)13m
- Solubility Rules16m
- Molecular Equations18m
- Types of Chemical Reactions12m
- Complete Ionic Equations18m
- Calculate Oxidation Numbers15m
- Redox Reactions17m
- Spontaneous Redox Reactions8m
- Balancing Redox Reactions: Acidic Solutions17m
- Balancing Redox Reactions: Basic Solutions17m
- Balancing Redox Reactions (Simplified)13m
- Galvanic Cell (Simplified)16m
- 6. Chemical Reactions & Quantities2h 35m
- 7. Energy, Rate and Equilibrium3h 46m
- Nature of Energy6m
- First Law of Thermodynamics7m
- Endothermic & Exothermic Reactions7m
- Bond Energy14m
- Thermochemical Equations12m
- Heat Capacity19m
- Thermal Equilibrium (Simplified)8m
- Hess's Law23m
- Rate of Reaction11m
- Energy Diagrams12m
- Chemical Equilibrium7m
- The Equilibrium Constant14m
- Le Chatelier's Principle23m
- Solubility Product Constant (Ksp)17m
- Spontaneous Reaction10m
- Entropy (Simplified)9m
- Gibbs Free Energy (Simplified)18m
- 8. Gases, Liquids and Solids3h 25m
- Pressure Units6m
- Kinetic Molecular Theory14m
- The Ideal Gas Law18m
- The Ideal Gas Law Derivations13m
- The Ideal Gas Law Applications6m
- Chemistry Gas Laws16m
- Chemistry Gas Laws: Combined Gas Law12m
- Standard Temperature and Pressure14m
- Dalton's Law: Partial Pressure (Simplified)13m
- Gas Stoichiometry18m
- Intermolecular Forces (Simplified)19m
- Intermolecular Forces and Physical Properties11m
- Atomic, Ionic and Molecular Solids10m
- Heating and Cooling Curves30m
- 9. Solutions4h 10m
- Solutions6m
- Solubility and Intermolecular Forces18m
- Solutions: Mass Percent6m
- Percent Concentrations10m
- Molarity18m
- Osmolarity15m
- Parts per Million (ppm)13m
- Solubility: Temperature Effect8m
- Intro to Henry's Law4m
- Henry's Law Calculations12m
- Dilutions12m
- Solution Stoichiometry14m
- Electrolytes (Simplified)13m
- Equivalents11m
- Molality15m
- The Colligative Properties15m
- Boiling Point Elevation16m
- Freezing Point Depression9m
- Osmosis16m
- Osmotic Pressure9m
- 10. Acids and Bases3h 29m
- Acid-Base Introduction11m
- Arrhenius Acid and Base6m
- Bronsted Lowry Acid and Base18m
- Acid and Base Strength17m
- Ka and Kb12m
- The pH Scale19m
- Auto-Ionization9m
- pH of Strong Acids and Bases9m
- Acid-Base Equivalents14m
- Acid-Base Reactions7m
- Gas Evolution Equations (Simplified)6m
- Ionic Salts (Simplified)23m
- Buffers25m
- Henderson-Hasselbalch Equation16m
- Strong Acid Strong Base Titrations (Simplified)10m
- 11. Nuclear Chemistry56m
- BONUS: Lab Techniques and Procedures1h 38m
- BONUS: Mathematical Operations and Functions47m
- 12. Introduction to Organic Chemistry1h 34m
- 13. Alkenes, Alkynes, and Aromatic Compounds2h 12m
- 14. Compounds with Oxygen or Sulfur1h 6m
- 15. Aldehydes and Ketones1h 1m
- 16. Carboxylic Acids and Their Derivatives1h 11m
- 17. Amines38m
- 18. Amino Acids and Proteins1h 51m
- 19. Enzymes1h 37m
- 20. Carbohydrates1h 46m
- Intro to Carbohydrates4m
- Classification of Carbohydrates4m
- Fischer Projections4m
- Enantiomers vs Diastereomers8m
- D vs L Enantiomers8m
- Cyclic Hemiacetals8m
- Intro to Haworth Projections4m
- Cyclic Structures of Monosaccharides11m
- Mutarotation4m
- Reduction of Monosaccharides10m
- Oxidation of Monosaccharides7m
- Glycosidic Linkage14m
- Disaccharides7m
- Polysaccharides7m
- 21. The Generation of Biochemical Energy2h 8m
- 22. Carbohydrate Metabolism2h 22m
- 23. Lipids2h 26m
- Intro to Lipids6m
- Fatty Acids25m
- Physical Properties of Fatty Acids6m
- Waxes4m
- Triacylglycerols12m
- Triacylglycerol Reactions: Hydrogenation8m
- Triacylglycerol Reactions: Hydrolysis13m
- Triacylglycerol Reactions: Oxidation7m
- Glycerophospholipids15m
- Sphingomyelins13m
- Steroids15m
- Cell Membranes7m
- Membrane Transport10m
- 24. Lipid Metabolism1h 45m
- 25. Protein and Amino Acid Metabolism1h 37m
- 26. Nucleic Acids and Protein Synthesis2h 54m
- Intro to Nucleic Acids4m
- Nitrogenous Bases16m
- Nucleoside and Nucleotide Formation9m
- Naming Nucleosides and Nucleotides13m
- Phosphodiester Bond Formation7m
- Primary Structure of Nucleic Acids11m
- Base Pairing10m
- DNA Double Helix6m
- Intro to DNA Replication20m
- Steps of DNA Replication11m
- Types of RNA10m
- Overview of Protein Synthesis4m
- Transcription: mRNA Synthesis9m
- Processing of pre-mRNA5m
- The Genetic Code6m
- Introduction to Translation7m
- Translation: Protein Synthesis18m
Intro to DNA Replication - Online Tutor, Practice Problems & Exam Prep
 Created using AI
Created using AIDNA replication begins with helicase unwinding the double helix at the origin of replication, forming replication forks. This process is semi-conservative, producing two new DNA strands, each containing one template and one daughter strand. The leading strand replicates continuously in the same direction as the fork, while the lagging strand forms discontinuous Okazaki fragments in the opposite direction. Key enzymes include helicase, stabilizing proteins, primase, DNA polymerase, and DNA ligase, which work together to ensure accurate replication and stability of the DNA structure.
Intro to DNA Replication Concept 1
Video transcript
Intro to DNA Replication Example 1
Video transcript
In this example question, it asks which of the following statements most accurately describes DNA replication:
A. It's semi-conservative, with all of the DNA copied and restructured within both new double helixes.
When we analyze the options provided, the terms mentioned include semi-conservative, dispersive, semi-conservative, and conservative. From our earlier discussion, we confirm that DNA replication follows a semi-conservative model, which narrows the correct answer to either option A or C. Options B and D are immediately eliminated.
To differentiate further between A and C:
- Option A states it is semi-conservative with all of the DNA copied and restructured within both new double helices.
- Option C states it is semi-conservative with the template strand being found within both double helices.
The correct description and more accurate answer is option C. This is because in the semi-conservative model of DNA replication, we replicate the template strand to create our complementary or daughter strand, resulting in two DNA double helices, each containing one template strand and one daughter strand. Option C correctly points out that the template strand will be found within both new double helices, making it the optimal choice. Hence, the final answer is C.
Intro to DNA Replication Concept 2
Video transcript
Now when we talk about DNA replication, we're going to say that it requires a host of multiple enzymes/proteins working together. Here, we've compiled the list of the most important enzymes and proteins necessary for DNA replication. First, we're going to start out with helicase. Helicase is an enzyme and will display it as a triangle that is yellow. Its job is to unwind the DNA double helix, and it does it at the replication fork. We'll talk about the replication fork later on as a kind of landmark that we find when we're talking about DNA replication. Now, when we say unwind, remember it's cutting through our hydrogen bonds between the nitrogenous bases in order to expose our template strands for replication.
Next, we have what are called stabilizing proteins. Here, these stabilizing proteins, we're going to depict them as these orange balls here. Their job is to bind to and stabilize the single-stranded DNA. Now, DNA wants to be in a double helix form. We need this protein to basically keep it open so that our replication can start. We can start to copy our template strands to produce our daughter strands.
Next, we have what's called our primase, which we're depicting as this image here, this pink magenta type color. Here, its job is to create RNA primers as a starting point for replication.
Next, we're going to have this bluish figure, this is our DNA Polymerase. Now, what is its job? Well, it creates a new DNA strand using the template strand of DNA. So, its job is to make our complementary or daughter strand. And then finally, we have DNA ligase. Here, what it does here is it's going to join, so it joins together new strands of DNA.
So again, it's all of these enzymes and proteins working in conjunction with one another that allows us to do DNA replication. Here we're just talking about what they resemble, what their names are, and what their function will be. Later on when we're talking about DNA replication, we'll see how the process actually happens. Alright. So keep in mind these important enzymes and proteins.
Intro to DNA Replication Concept 3
Video transcript
Hey everyone. So in our continued discussion of DNA replication, we can say that replication begins with helicase unwinding DNA at a specific site called the origin of replication. Now, this origin of replication or ori, this is basically where helicase decides where it wants to cut into the hydrogen bonds between our nitrogenous bases. This typically happens when it's scanning for a specific sequence of nucleotides, and that's where it starts to cut.
So, if we take a look here, we'd say that this orange box that is on top of this Phosphate Sugar Backbone of our DNA Double Helix represents our origin of replication. You can see here we have a helicase and it looks like it's moving this way, and another helicase that's moving this way. They are bending and then breaking these hydrogen bonds that are connecting the different nitrogenous bases together. Now, here we're going to say that our two strands of DNA are separated, forming what we call 2 replication forks. These replication forks are Y-shaped regions at each end of the bubble where DNA is unwound.
Now, if I take a look here, we still have our origin of replication here, we have our helicase here and our helicase here. They're just continually cutting through the hydrogen bonds, opening up our DNA Double Helix where we have this gap now. This empty space in here represents our replication bubble. And we're going to say that the regions that are highlighted in yellow, well, those are our replication forks. Remember, we say they are Y-shaped regions. If you look, it kind of looks like a Y. Looks like a Y here too. So they are Y-shaped regions.
Now, DNA replication proceeds bidirectionally in both directions. So, as you can see here, we have our old DNA in blue and then we have our new DNA being laid down once we've opened up the double helix. We can see that DNA is being laid down in this direction and DNA is being laid down in this direction. We also see that we have a magenta tail, that's our RNA primer, which remember, RNA primers are put down by our enzyme, primase, that first has to be put down before our new DNA can start being put down, complementary to the DNA template strands.
Alright. So just remember, when we're talking about replication forks, we're talking about this region highlighted in yellow, looks like a Y. It's being created by helicase just continuously moving down the double helix and cutting through the hydrogen bonds that connect the nitrogenous bases together. Opening it up creates a replication bubble which then allows new DNA to come in and start being laid down on the DNA template strands. With this new DNA, we have DNA replication.
Intro to DNA Replication Example 2
Video transcript
Which of the following is incorrect regarding DNA replication forks? DNA replication forks begin forming at the origin of replication, also known as the ori. That's true. Helicase comes there and then they start cutting through, and we're starting to form replication forks right then and there. DNA replication forks are caused by helicase separating two complementary strands of DNA. That is true. The two strands of DNA are complementary to each other. They run antiparallel to each other. So this is true. There is one replication fork found for every replication bubble formed. So remember, we have our replication bubble. Alright. We have our replication bubble that's formed. We know that this region here and this region here represent our replication forks. And we know we have our origin of replication there. From this image, we can see that one replication bubble is associated with two replication forks. So this statement is incorrect. It's not one replication fork for every replication bubble. It's two replication forks for every one replication bubble. Now, DNA replication forks are found at both ends of the replication bubble. We can see from this drawing that this is true as well. So here, the only statement that's incorrect is option C. Remember, one replication bubble will have two replication forks, one at each end of the bubble.
How many helicase enzymes are needed for a strand of DNA that possess 5 origins of replication?
2
5
6
8
10
Intro to DNA Replication Concept 4
Video transcript
In this video, we continue our discussion of DNA replication in terms of the lagging strand and the leading strand. Now, here, after separating the double helix, replication creates 2 new strands. We're going to say the strand types are dependent on the direction of the replication fork. Here we have our leading strand and our lagging strand. We're going to say the leading strand is the continuous replication in the same direction as the replication fork movement, and only 1 RNA primer is required for replication. If we come down here and take a look at this image, remember, we have our old DNA in blue, our new DNA being laid down in green, and then our RNA primer that's being put down by primases to help initiate replication. Now, imagine that we have our origin of replication, and it's over here. Everything that's going on is to the left of that right now. We're just paying attention to half of our replication bubble.
If we take a look at this, we're going to look at our leading strand first. We say that we put down our primer and remember, when putting down our primer and starting to create new DNA, it always goes in the 5' to 3' direction. So here, this is the 3' end of our old DNA. If we're going from 5' to 3', that means we have to lay down our initial primer and then the new DNA on the 5' end of itself. And it's going to go this way towards 3' for itself. 3' for itself would correlate to 5' of the old DNA strand. Now, we have our helicase here which is cutting through the hydrogen bonds of our nitrogenous bases. So this is our replication fork here. Our helicase is moving in this general direction, and so our replication fork is moving in that same direction. The leading strand puts down new DNA in that same direction. It's moving this way, in the same direction as the helicase and the replication fork. So as this is becoming more and more open, we're putting more and more DNA down.
The lagging strand though is the opposite. Here, it is a discontinuous replication in the opposite direction as the replication fork movement. Here we're going to create what are called Okazaki fragments, which are multiple, small replicated segments that require an RNA primer. So if we take a look here, we're going to say that an Okazaki fragment is just a small segment of new DNA coupled with our primer that's been put down. So we can see that we have 1, 2, 3, 4, 5 Okazaki fragments shown here. Collectively altogether, this is what is the lagging strand. So our lagging strand is just a collection of Okazaki fragments.
Now, how does this work? Well, our replication fork and our helicase are going in this direction. We see that new DNA is being put down in this direction. It's going the opposite way. This indicates a lagging strand. Now, the way it works is as we're opening up this DNA helix with our replication bubble being formed we're putting down a primer. So, a primer gets laid down here as we open up and then new DNA starts getting created. But then what happens? This mechanism keeps sliding down this way, opening up the DNA strand even more. And as it opens up even more, then another primer has to be put down here and new DNA moves forward. So, we're basically waiting for it to open up so we could put a primer there and then grow this way. It opens up a little bit further down here, we put another primer, it moves down that way. We're waiting for it to open up even more the DNA double helix, so we can put a primer and then move in the opposite direction. This is going to cause small gaps in between our Okazaki fragments. Later on, we'll see how we can connect these gaps together at the end of DNA replication. But for now, our lagging strand, it puts down new DNA in the opposite direction of the replication fork movement. This creates these Okazaki fragments and they have small gaps between them. This is different from the leading strand which just continuously puts more and more DNA down as long as it needs to to make a whole strand of new DNA. It's moving in the same direction as the helicase and the replication fork movement. We don't have these small gaps that we would see in the lagging strand when it comes to the leading strand.
Alright. So just keep in mind, the difference between our leading strand and our lagging strand is really in reference to the direction of our replication fork movement and the helicase enzyme.
Intro to DNA Replication Example 3
Video transcript
Here we're told that a newly synthesized leading strand of DNA is given as follows: from the 3' end, we have ATTCGACTAA towards the 5' end. It asks to determine the template DNA strand that was copied. Remember, this leading strand was copied from our template strand which is our original or parent strand. Here, they need to be antiparallel to one another. So this end will be 5', and this end here will be 3'. And remember, this is DNA, so the base pairings are A with T and G with C. So here, this is A, so this originally had to be T. This was A and A. This is C, so this was a G. This is a G, so this was a C. This is a T. This is a G. This is an A, and then this is T and T. So this was our original template DNA strand that was copied to make this leading strand. So this would be our final answer.
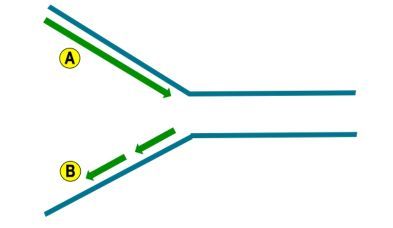
Based on the image below, which of the following statements is true?
Arrow A represents the lagging strand and moves in the opposite direction of the replication fork movement.
Arrow B represents the lagging strand and moves in the same direction of the replication fork movement.
Arrow A represents the leading strand and moves in the same direction of the replication fork movement.
Arrow B represents the leading strand and moves in the opposite direction of the replication fork movement.
Do you want more practice?
Here’s what students ask on this topic:
What is the semi-conservative model of DNA replication?
The semi-conservative model of DNA replication describes how each new DNA molecule consists of one original (template) strand and one newly synthesized (daughter) strand. During replication, the double helix unwinds, and each template strand serves as a guide for creating a complementary daughter strand. This model ensures that each daughter DNA molecule retains half of the original DNA, preserving genetic information across generations. The process involves key enzymes like helicase, which unwinds the DNA, and DNA polymerase, which synthesizes the new strands. This method of replication helps maintain the integrity and stability of the genetic code.
 Created using AI
Created using AIWhat are the key enzymes involved in DNA replication?
DNA replication involves several key enzymes and proteins working together to ensure accurate and efficient replication. The main enzymes include:
- Helicase: Unwinds the DNA double helix at the replication fork.
- Stabilizing Proteins: Bind to single-stranded DNA to keep it open for replication.
- Primase: Synthesizes RNA primers to provide a starting point for DNA synthesis.
- DNA Polymerase: Adds nucleotides to the growing DNA strand, using the template strand as a guide.
- DNA Ligase: Joins Okazaki fragments on the lagging strand, sealing gaps to create a continuous DNA strand.
These enzymes work in concert to ensure the DNA is accurately copied and the genetic information is preserved.
 Created using AI
Created using AIWhat is the difference between the leading and lagging strands in DNA replication?
During DNA replication, the leading and lagging strands are synthesized differently due to the antiparallel nature of DNA. The leading strand is synthesized continuously in the same direction as the replication fork movement, requiring only one RNA primer. In contrast, the lagging strand is synthesized discontinuously in the opposite direction of the replication fork, forming short segments called Okazaki fragments. Each fragment requires a new RNA primer. DNA ligase later joins these fragments to form a continuous strand. This difference arises because DNA polymerase can only add nucleotides in the 5' to 3' direction.
 Created using AI
Created using AIWhat is the role of helicase in DNA replication?
Helicase plays a crucial role in DNA replication by unwinding the double helix at the origin of replication. It breaks the hydrogen bonds between the nitrogenous bases, creating two single-stranded DNA templates. This unwinding forms Y-shaped structures called replication forks, where the DNA is separated and replication can proceed. By exposing the template strands, helicase allows other enzymes, such as DNA polymerase, to synthesize new complementary strands. Without helicase, the DNA double helix would remain intact, preventing the replication process from occurring.
 Created using AI
Created using AIWhat are Okazaki fragments and how are they formed?
Okazaki fragments are short segments of DNA synthesized on the lagging strand during DNA replication. They are formed because DNA polymerase can only add nucleotides in the 5' to 3' direction, while the lagging strand runs in the 3' to 5' direction relative to the replication fork. As the replication fork opens, primase lays down RNA primers at intervals, and DNA polymerase synthesizes short DNA segments between these primers. These segments, called Okazaki fragments, are later joined by DNA ligase to form a continuous DNA strand. This process ensures that the entire lagging strand is accurately replicated.
 Created using AI
Created using AI