Here are the essential concepts you must grasp in order to answer the question correctly.
Base Pair
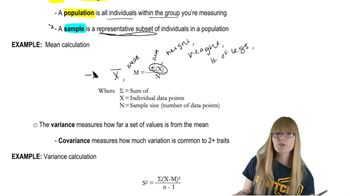
A base pair consists of two nucleotides on opposite complementary DNA or RNA strands that are connected via hydrogen bonds. In DNA, the base pairs are adenine-thymine (A-T) and guanine-cytosine (G-C). Understanding base pairs is crucial for calculating the total number of pairs in a given length of DNA.
Recommended video:
DNA Length Measurement
DNA length can be measured in micrometers (µm) or base pairs. One base pair of DNA is approximately 0.34 nanometers long. To convert the length of DNA from micrometers to base pairs, one must use this conversion factor, which allows for the determination of the total number of base pairs in a given length of DNA.
Recommended video:
Mathematical Measurements
Phage T2
Phage T2 is a type of bacteriophage, a virus that infects bacteria, specifically Escherichia coli. Its DNA is a double-stranded molecule that serves as a model for studying viral genetics. Understanding the structure and properties of phage T2 DNA is essential for answering questions related to its length and base pair composition.
Recommended video:
 Verified step by step guidance
Verified step by step guidance