Much of what we know about gene interactions in development has been learned using nematodes, yeast, flies, and bacteria. This is due, in part, to the relative ease of genetic manipulation of these well-characterized genomes. However, of great interest are gene interactions involving complex diseases in humans. Wang and White [(2011). Nature Methods 8(4):341–346] describe work using RNAi to examine the interactive proteome in mammalian cells. They mention that knockdown inefficiencies and off-target effects of introduced RNAi species are areas that need particular improvement if the methodology is to be fruitful.
Comment on how 'knockdown inefficiencies' and 'off-target effects' would influence the interpretation of results.
Regulation of the lac operon in E. coli (see Chapter 16) and regulation of the GAL system in yeast are analogous in that they both serve to adapt cells to growth on different carbon sources. However, the transcriptional changes are accomplished very differently. Consider the conceptual similarities and differences as you address the following.
Compare and contrast the cis-regulatory elements of the lac operon and GAL gene system.
 Verified step by step guidance
Verified step by step guidance
Verified Solution
Key Concepts
Cis-Regulatory Elements
Lac Operon
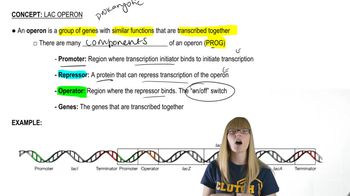
GAL Gene System

In this chapter, we discussed several specific cis-elements in mRNAs that regulate splicing, stability, decay, localization, and translation. However, it is likely that many other uncharacterized cis-elements exist. One way in which they may be characterized is through the use of a reporter gene such as the gene encoding the green fluorescent protein (GFP) from jellyfish. GFP emits green fluorescence when excited by blue light. Explain how one might be able to devise an assay to test for the effect of various cis-elements on posttranscriptional gene regulation using cells that transcribe a GFP mRNA with genetically inserted cis-elements.
Regulation of the lac operon in E. coli (see Chapter 16) and regulation of the GAL system in yeast are analogous in that they both serve to adapt cells to growth on different carbon sources. However, the transcriptional changes are accomplished very differently. Consider the conceptual similarities and differences as you address the following.
Compare and contrast the roles of the lac operon inducer in bacteria and Gal3p in eukaryotes in the regulation of their respective systems.
Regulation of the lac operon in E. coli (see Chapter 16) and regulation of the GAL system in yeast are analogous in that they both serve to adapt cells to growth on different carbon sources. However, the transcriptional changes are accomplished very differently. Consider the conceptual similarities and differences as you address the following.
Compare and contrast how these two systems are negatively regulated such that they are downregulated in the presence of glucose.
Incorrectly spliced RNAs often lead to human pathologies. Scientists have examined cancer cells for splice-specific changes and found that many of the changes disrupt tumor-suppressor gene function [Xu and Lee (2003). Nucl. Acids Res. 31:5635–5643]. In general, what would be the effects of splicing changes on these RNAs and the function of tumor-suppressor gene function? How might loss of splicing specificity be associated with cancer?
Mutations in the low-density lipoprotein receptor (LDLR) gene are a primary cause of familial hypercholesterolemia. One such mutation is a SNP in exon 12 of the LDLR. In premenopausal women, but not in men or postmenopausal women, this SNP leads to skipping of exon 12 and production of a truncated nonfunctional protein. It is hypothesized that this SNP compromises a splice enhancer [Zhu et al. (2007). Hum Mol Genet. 16:1765–1772]. What are some possible ways in which this SNP can lead to this defect, but only in premenopausal women?
