In this video, we're going to talk about how to determine the predominant species of a molecule. So in our last lesson video, we reviewed the Henderson Hasselbalch equation from your previous chemistry courses, and we know that the Henderson Hasselbalch equation reveals the ratio of the concentration of the conjugate base to the concentration of the conjugate acid. And it turns out that the conjugate base and the conjugate acid are actually different forms or species of a molecule. And so the predominant species is really just referring to the most abundant form of a molecule that exists under a specific set of conditions. And so it turns out that the pH of the solution, as well as the pKa of the acid, will determine or dictate the predominant species of that acidic molecule. And so in our next lesson video, we're going to compare the pH of the solution and the pKas of acids to determine the predominant species of those acidic molecules. So I'll see you guys in that lesson video.
- 1. Introduction to Biochemistry4h 34m
- What is Biochemistry?5m
- Characteristics of Life12m
- Abiogenesis13m
- Nucleic Acids16m
- Proteins12m
- Carbohydrates8m
- Lipids10m
- Taxonomy10m
- Cell Organelles12m
- Endosymbiotic Theory11m
- Central Dogma22m
- Functional Groups15m
- Chemical Bonds13m
- Organic Chemistry31m
- Entropy17m
- Second Law of Thermodynamics11m
- Equilibrium Constant10m
- Gibbs Free Energy37m
- 2. Water3h 23m
- 3. Amino Acids8h 10m
- Amino Acid Groups8m
- Amino Acid Three Letter Code13m
- Amino Acid One Letter Code37m
- Amino Acid Configuration20m
- Essential Amino Acids14m
- Nonpolar Amino Acids21m
- Aromatic Amino Acids14m
- Polar Amino Acids16m
- Charged Amino Acids40m
- How to Memorize Amino Acids1h 7m
- Zwitterion33m
- Non-Ionizable Vs. Ionizable R-Groups11m
- Isoelectric Point10m
- Isoelectric Point of Amino Acids with Ionizable R-Groups51m
- Titrations of Amino Acids with Non-Ionizable R-Groups44m
- Titrations of Amino Acids with Ionizable R-Groups38m
- Amino Acids and Henderson-Hasselbalch44m
- 4. Protein Structure10h 4m
- Peptide Bond18m
- Primary Structure of Protein31m
- Altering Primary Protein Structure15m
- Drawing a Peptide44m
- Determining Net Charge of a Peptide42m
- Isoelectric Point of a Peptide37m
- Approximating Protein Mass7m
- Peptide Group22m
- Ramachandran Plot26m
- Atypical Ramachandran Plots12m
- Alpha Helix15m
- Alpha Helix Pitch and Rise20m
- Alpha Helix Hydrogen Bonding24m
- Alpha Helix Disruption23m
- Beta Strand12m
- Beta Sheet12m
- Antiparallel and Parallel Beta Sheets39m
- Beta Turns26m
- Tertiary Structure of Protein16m
- Protein Motifs and Domains23m
- Denaturation14m
- Anfinsen Experiment20m
- Protein Folding34m
- Chaperone Proteins19m
- Prions4m
- Quaternary Structure15m
- Simple Vs. Conjugated Proteins10m
- Fibrous and Globular Proteins11m
- 5. Protein Techniques14h 5m
- Protein Purification7m
- Protein Extraction5m
- Differential Centrifugation15m
- Salting Out18m
- Dialysis9m
- Column Chromatography11m
- Ion-Exchange Chromatography35m
- Anion-Exchange Chromatography38m
- Size Exclusion Chromatography28m
- Affinity Chromatography16m
- Specific Activity16m
- HPLC29m
- Spectrophotometry51m
- Native Gel Electrophoresis23m
- SDS-PAGE34m
- SDS-PAGE Strategies16m
- Isoelectric Focusing17m
- 2D-Electrophoresis23m
- Diagonal Electrophoresis29m
- Mass Spectrometry12m
- Mass Spectrum47m
- Tandem Mass Spectrometry16m
- Peptide Mass Fingerprinting16m
- Overview of Direct Protein Sequencing30m
- Amino Acid Hydrolysis10m
- FDNB26m
- Chemical Cleavage of Bonds29m
- Peptidases1h 6m
- Edman Degradation30m
- Edman Degradation Sequenator and Sequencing Data Analysis4m
- Edman Degradation Reaction Efficiency20m
- Ordering Cleaved Fragments21m
- Strategy for Ordering Cleaved Fragments58m
- Indirect Protein Sequencing Via Geneomic Analyses24m
- 6. Enzymes and Enzyme Kinetics13h 38m
- Enzymes24m
- Enzyme-Substrate Complex17m
- Lock and Key Vs. Induced Fit Models23m
- Optimal Enzyme Conditions9m
- Activation Energy24m
- Types of Enzymes41m
- Cofactor15m
- Catalysis19m
- Electrostatic and Metal Ion Catalysis11m
- Covalent Catalysis18m
- Reaction Rate10m
- Enzyme Kinetics24m
- Rate Constants and Rate Law35m
- Reaction Orders52m
- Rate Constant Units11m
- Initial Velocity31m
- Vmax Enzyme27m
- Km Enzyme42m
- Steady-State Conditions25m
- Michaelis-Menten Assumptions18m
- Michaelis-Menten Equation52m
- Lineweaver-Burk Plot43m
- Michaelis-Menten vs. Lineweaver-Burk Plots20m
- Shifting Lineweaver-Burk Plots37m
- Calculating Vmax40m
- Calculating Km31m
- Kcat46m
- Specificity Constant1h 1m
- 7. Enzyme Inhibition and Regulation 8h 42m
- Enzyme Inhibition13m
- Irreversible Inhibition12m
- Reversible Inhibition9m
- Inhibition Constant26m
- Degree of Inhibition15m
- Apparent Km and Vmax29m
- Inhibition Effects on Reaction Rate10m
- Competitive Inhibition52m
- Uncompetitive Inhibition33m
- Mixed Inhibition40m
- Noncompetitive Inhibition26m
- Recap of Reversible Inhibition37m
- Allosteric Regulation7m
- Allosteric Kinetics17m
- Allosteric Enzyme Conformations33m
- Allosteric Effectors18m
- Concerted (MWC) Model25m
- Sequential (KNF) Model20m
- Negative Feedback13m
- Positive Feedback15m
- Post Translational Modification14m
- Ubiquitination19m
- Phosphorylation16m
- Zymogens13m
- 8. Protein Function 9h 41m
- Introduction to Protein-Ligand Interactions15m
- Protein-Ligand Equilibrium Constants22m
- Protein-Ligand Fractional Saturation32m
- Myoglobin vs. Hemoglobin27m
- Heme Prosthetic Group31m
- Hemoglobin Cooperativity23m
- Hill Equation21m
- Hill Plot42m
- Hemoglobin Binding in Tissues & Lungs31m
- Hemoglobin Carbonation & Protonation19m
- Bohr Effect23m
- BPG Regulation of Hemoglobin24m
- Fetal Hemoglobin6m
- Sickle Cell Anemia24m
- Chymotrypsin18m
- Chymotrypsin's Catalytic Mechanism38m
- Glycogen Phosphorylase21m
- Liver vs Muscle Glycogen Phosphorylase21m
- Antibody35m
- ELISA15m
- Motor Proteins14m
- Skeletal Muscle Anatomy22m
- Skeletal Muscle Contraction45m
- 9. Carbohydrates7h 49m
- Carbohydrates19m
- Monosaccharides15m
- Stereochemistry of Monosaccharides33m
- Monosaccharide Configurations32m
- Cyclic Monosaccharides20m
- Hemiacetal vs. Hemiketal19m
- Anomer14m
- Mutarotation13m
- Pyranose Conformations23m
- Common Monosaccharides33m
- Derivatives of Monosaccharides21m
- Reducing Sugars21m
- Reducing Sugars Tests19m
- Glycosidic Bond48m
- Disaccharides40m
- Glycoconjugates12m
- Polysaccharide7m
- Cellulose7m
- Chitin8m
- Peptidoglycan12m
- Starch13m
- Glycogen14m
- Lectins16m
- 10. Lipids5h 49m
- Lipids15m
- Fatty Acids30m
- Fatty Acid Nomenclature11m
- Omega-3 Fatty Acids12m
- Triacylglycerols11m
- Glycerophospholipids24m
- Sphingolipids13m
- Sphingophospholipids8m
- Sphingoglycolipids12m
- Sphingolipid Recap22m
- Waxes5m
- Eicosanoids19m
- Isoprenoids9m
- Steroids14m
- Steroid Hormones11m
- Lipid Vitamins19m
- Comprehensive Final Lipid Map13m
- Biological Membranes16m
- Physical Properties of Biological Membranes18m
- Types of Membrane Proteins8m
- Integral Membrane Proteins16m
- Peripheral Membrane Proteins12m
- Lipid-Linked Membrane Proteins21m
- 11. Biological Membranes and Transport 6h 37m
- Biological Membrane Transport21m
- Passive vs. Active Transport18m
- Passive Membrane Transport21m
- Facilitated Diffusion8m
- Erythrocyte Facilitated Transporter Models30m
- Membrane Transport of Ions29m
- Primary Active Membrane Transport15m
- Sodium-Potassium Ion Pump20m
- SERCA: Calcium Ion Pump10m
- ABC Transporters12m
- Secondary Active Membrane Transport12m
- Glucose Active Symporter Model19m
- Endocytosis & Exocytosis18m
- Neurotransmitter Release23m
- Summary of Membrane Transport21m
- Thermodynamics of Membrane Diffusion: Uncharged Molecule51m
- Thermodynamics of Membrane Diffusion: Charged Ion1h 1m
- 12. Biosignaling9h 45m
- Introduction to Biosignaling44m
- G protein-Coupled Receptors32m
- Stimulatory Adenylate Cyclase GPCR Signaling42m
- cAMP & PKA28m
- Inhibitory Adenylate Cyclase GPCR Signaling29m
- Drugs & Toxins Affecting GPCR Signaling20m
- Recap of Adenylate Cyclase GPCR Signaling5m
- Phosphoinositide GPCR Signaling58m
- PSP Secondary Messengers & PKC27m
- Recap of Phosphoinositide Signaling7m
- Receptor Tyrosine Kinases26m
- Insulin28m
- Insulin Receptor23m
- Insulin Signaling on Glucose Metabolism57m
- Recap Of Insulin Signaling in Glucose Metabolism6m
- Insulin Signaling as a Growth Factor1h 1m
- Recap of Insulin Signaling As A Growth Factor9m
- Recap of Insulin Signaling1m
- Jak-Stat Signaling25m
- Lipid Hormone Signaling15m
- Summary of Biosignaling13m
- Signaling Defects & Cancer20m
- Review 1: Nucleic Acids, Lipids, & Membranes2h 47m
- Nucleic Acids 19m
- Nucleic Acids 211m
- Nucleic Acids 34m
- Nucleic Acids 44m
- DNA Sequencing 19m
- DNA Sequencing 211m
- Lipids 111m
- Lipids 24m
- Membrane Structure 110m
- Membrane Structure 29m
- Membrane Transport 18m
- Membrane Transport 24m
- Membrane Transport 36m
- Practice - Nucleic Acids 111m
- Practice - Nucleic Acids 23m
- Practice - Nucleic Acids 39m
- Lipids11m
- Practice - Membrane Structure 17m
- Practice - Membrane Structure 25m
- Practice - Membrane Transport 16m
- Practice - Membrane Transport 26m
- Review 2: Biosignaling, Glycolysis, Gluconeogenesis, & PP-Pathway3h 12m
- Biosignaling 19m
- Biosignaling 219m
- Biosignaling 311m
- Biosignaling 49m
- Glycolysis 17m
- Glycolysis 27m
- Glycolysis 38m
- Glycolysis 410m
- Fermentation6m
- Gluconeogenesis 18m
- Gluconeogenesis 27m
- Pentose Phosphate Pathway15m
- Practice - Biosignaling13m
- Practice - Bioenergetics 110m
- Practice - Bioenergetics 216m
- Practice - Glycolysis 111m
- Practice - Glycolysis 27m
- Practice - Gluconeogenesis5m
- Practice - Pentose Phosphate Path6m
- Review 3: Pyruvate & Fatty Acid Oxidation, Citric Acid Cycle, & Glycogen Metabolism2h 26m
- Pyruvate Oxidation9m
- Citric Acid Cycle 114m
- Citric Acid Cycle 27m
- Citric Acid Cycle 37m
- Citric Acid Cycle 411m
- Metabolic Regulation 18m
- Metabolic Regulation 213m
- Glycogen Metabolism 16m
- Glycogen Metabolism 28m
- Fatty Acid Oxidation 111m
- Fatty Acid Oxidation 28m
- Citric Acid Cycle Practice 17m
- Citric Acid Cycle Practice 26m
- Citric Acid Cycle Practice 32m
- Glucose and Glycogen Regulation Practice 14m
- Glucose and Glycogen Regulation Practice 26m
- Fatty Acid Oxidation Practice 14m
- Fatty Acid Oxidation Practice 27m
- Review 4: Amino Acid Oxidation, Oxidative Phosphorylation, & Photophosphorylation1h 48m
- Amino Acid Oxidation 15m
- Amino Acid Oxidation 211m
- Oxidative Phosphorylation 18m
- Oxidative Phosphorylation 210m
- Oxidative Phosphorylation 310m
- Oxidative Phosphorylation 47m
- Photophosphorylation 15m
- Photophosphorylation 29m
- Photophosphorylation 310m
- Practice: Amino Acid Oxidation 12m
- Practice: Amino Acid Oxidation 22m
- Practice: Oxidative Phosphorylation 15m
- Practice: Oxidative Phosphorylation 24m
- Practice: Oxidative Phosphorylation 35m
- Practice: Photophosphorylation 15m
- Practice: Photophosphorylation 21m
Determining Predominate Species - Online Tutor, Practice Problems & Exam Prep
 Created using AI
Created using AIDetermining Predominate Species
Video transcript
Determining Predominate Species
Video transcript
So we can determine the predominant species of a molecule by comparing the solution pH to an acid's pKa, which will reveal the relative concentrations of the conjugate base, as well as the concentration of the conjugate acid. Recall that the conjugate bases are deprotonated molecules because they have one less hydrogen than the conjugate acid. The conjugate acids, of course, are going to be protonated molecules, and that's because they have one more hydrogen than the conjugate base. In the example below, we're going to compare the pH and the pKa to fill in all of the blanks in our chart below. When we say compare the pH to the pKa, we mean to compare the value of the pH to the value of the pKa and determine if those two values are either equal to each other, or if one value is greater than or less than the other. One way that helps me determine the predominant species is by always associating the 'a' in the pKa to the 'a' in the concentration of conjugate acid. By always associating the pKa with the concentration of conjugate acid, I'm able to quickly determine the predominant species. Let me show you what I mean.
When the pH is equal to the pKa, I know that the concentration of conjugate base CBCA is going to be equal to the concentration of conjugate acid. Pretty easy, right? When the pH is less than the pKa, because I'm associating the pKa with the concentration of conjugate acid, I know that the concentration of conjugate base is going to be less than the concentration of conjugate acid. Essentially, it's going to have the same relationship as the pH over pKa. And so, when the pH is greater than the pKa, again, because I'm associating pKa with the concentration of conjugate acid, I know that the concentration of conjugate base will be greater than the concentration of conjugate acid. That's pretty easy, right? Associate the pKa with the conjugate acid, and you'll be able to quickly determine the predominant species. The predominant species will be the one in the highest abundance.
Notice when the pH is equal to the pKa, the concentration of conjugate base is exactly equal to the concentration of conjugate acid, which means that there is no predominant species since neither is in the highest abundance. They're both equal to each other, and that means that 50% of our molecules are going to be in a deprotonated conjugate base form, and the other 50% of our molecules are going to be in a protonated conjugate acid form. When the pH is less than the pKa, the conjugate acid is in higher concentration, which means the conjugate acid is predominating. We know that the majority of the molecules are going to be protonated, since our conjugate acid is always protonated. However, when the pH is greater than the pKa, the conjugate base is in higher concentration so the conjugate base predominates. We know that the conjugate base is a deprotonated form, so that means that the majority of the molecules are going to be deprotonated.
In our final column over here on the far right, notice we have the Henderson-Hasselbalch equation, which we know is pH=pKa+logCBCA. You'll notice here that we have the same exact equation for all three of these different scenarios, and the only thing that's going to be different is this ratio of the concentration of conjugate base to the concentration of conjugate acid. When the pH is equal to the pKa, we know that the concentration of conjugate base, which I'll abbreviate with CB, is going to be relatively equal to the ratios of the conjugate acid, abbreviated CA. When the pH is less than the pKa, we know that the conjugate acid is predominating and most of the molecules are going to be protonated. So, that means that we're going to have a large concentration of conjugate acid, and on the other hand, we're going to have a much smaller concentration of conjugate base. With this last scenario, when the pH is greater than the pKa, because we know that the conjugate base is going to be predominating and most of the molecules are going to be deprotonated, indicating that we have a large concentration of conjugate base, and a smaller concentration of conjugate acid. These sizes represent what we see in this column over here. Moving forward, we'll utilize a lot of these concepts, and it's important for you to take some time to study this chart and to commit some of it to your understanding. Rewatch this video a couple of times, and we'll get some practice moving forward in our next couple of practice videos. I'll see you guys there.
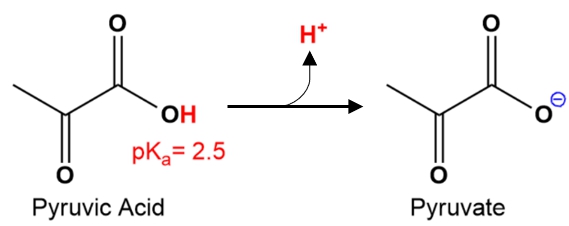
Consider the following pKa value for pyruvic acid. Which of the following species predominates at pH = 7.4?
Here’s what students ask on this topic:
What is the Henderson-Hasselbalch equation and how is it used to determine the predominant species of a molecule?
The Henderson-Hasselbalch equation is used to relate the pH of a solution to the pKa of an acid and the ratio of the concentrations of its conjugate base and conjugate acid. The equation is:
To determine the predominant species, compare the solution pH to the acid's pKa. If pH = pKa, the concentrations of conjugate acid and base are equal. If pH < pKa, the conjugate acid predominates. If pH > pKa, the conjugate base predominates. This helps in understanding the acid-base equilibria in the solution.
 Created using AI
Created using AIHow do you determine the predominant species when the pH is equal to the pKa?
When the pH of a solution is equal to the pKa of the acid, the concentrations of the conjugate acid and conjugate base are equal. This means that neither species predominates, and the solution contains 50% conjugate acid and 50% conjugate base. This equilibrium state is crucial for buffer solutions, which resist changes in pH upon the addition of small amounts of acid or base.
 Created using AI
Created using AIWhat happens to the predominant species when the pH is less than the pKa?
When the pH of a solution is less than the pKa of the acid, the conjugate acid predominates. This is because the lower pH indicates a higher concentration of hydrogen ions (H+), which shifts the equilibrium towards the protonated form of the molecule. Therefore, the majority of the molecules will be in the conjugate acid form, which is protonated.
 Created using AI
Created using AIWhat is the relationship between pH, pKa, and the concentrations of conjugate acid and base?
The relationship between pH, pKa, and the concentrations of conjugate acid and base is described by the Henderson-Hasselbalch equation:
When pH = pKa, the concentrations of conjugate acid and base are equal. If pH < pKa, the conjugate acid predominates, and if pH > pKa, the conjugate base predominates. This equation helps predict the predominant species in a solution based on its pH and the acid's pKa.
 Created using AI
Created using AIWhy is it important to understand the concept of predominant species in biochemistry?
Understanding the concept of predominant species is crucial in biochemistry because it helps predict the behavior of molecules in different pH environments. This knowledge is essential for enzyme activity, drug design, and metabolic pathways, as the ionization state of molecules can affect their function, solubility, and interaction with other molecules. By knowing the predominant species, biochemists can better understand and manipulate biochemical processes.
 Created using AI
Created using AI