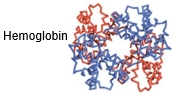
Building on the understanding of protein structure, we now explore the quaternary protein structure, which represents the highest level of organization in proteins. Quaternary structure involves the assembly of multiple polypeptide chains, known as subunits, into a single protein complex. Each subunit can be identical, referred to as homo, or different, termed hetero.
In terms of nomenclature, the number of subunits in a protein complex is indicated by specific terms: dimers consist of two subunits, trimers consist of three, and tetramers consist of four. For example, a dimer can be classified as a homodimer if both subunits are identical, while it is a heterodimer if the subunits differ. Similarly, a trimer with three distinct subunits is called a heterotrimer, and a tetramer with four different subunits is labeled a heterotetramer. The distinction between homo and hetero is crucial; if any subunit differs, the entire complex is classified as hetero.
This understanding of quaternary structure is essential for grasping how proteins function in biological systems, as the arrangement and interaction of subunits can significantly influence a protein's activity and stability. As we continue to study protein structures, these concepts will be applied in practical scenarios to reinforce learning.