How do geneticists detect the presence of genetic variation as different alleles in a population?

How do we know the age of the last common ancestor shared by two species?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
Molecular Clock

Phylogenetic Trees
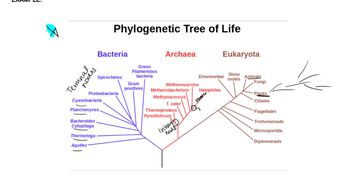
Fossil Record and Calibration
How do we know whether the genetic structure of a population is static or dynamic?
How do we know when populations have diverged to the point that they form two different species?
Write a short essay describing the roles of mutation, migration, and selection in bringing about speciation.
Price et al. [(1999). J. Bacteriol. 181:2358–2362] conducted a genetic study of the toxin transport protein (PA) of Bacillus anthracis, the bacterium that causes anthrax in humans. Within the 2294-nucleotide gene in 26 strains they identified five point mutations—two missense and three synonyms—among different isolates. Necropsy samples from an anthrax outbreak in 1979 revealed a novel missense mutation and five unique nucleotide changes among ten victims. The authors concluded that these data indicate little or no horizontal transfer between different B. anthracis strains.
Which types of nucleotide changes (missense or synonyms) cause amino acid changes?
Price et al. [(1999). J. Bacteriol. 181:2358–2362] conducted a genetic study of the toxin transport protein (PA) of Bacillus anthracis, the bacterium that causes anthrax in humans. Within the 2294-nucleotide gene in 26 strains they identified five point mutations—two missense and three synonyms—among different isolates. Necropsy samples from an anthrax outbreak in 1979 revealed a novel missense mutation and five unique nucleotide changes among ten victims. The authors concluded that these data indicate little or no horizontal transfer between different B. anthracis strains.
What is meant by 'horizontal transfer'?
