It has been noted that most transposons in humans and other organisms are located in noncoding regions of the genome—regions such as introns, pseudogenes, and stretches of particular types of repetitive DNA. There are several ways to interpret this observation. Describe two possible interpretations. Which interpretation do you favor? Why?

Verified Solution
Key Concepts
Transposons
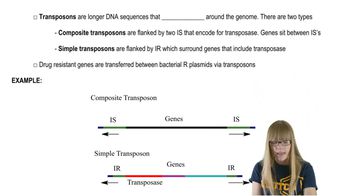
Noncoding DNA

Genomic Evolution

Shown here are the amino acid sequences of the wild-type and three mutant forms of a short protein.
___________________________________________________
Wild-type: Met-Trp-Tyr-Arg-Gly-Ser-Pro-Thr
Mutant 1: Met-Trp
Mutant 2: Met-Trp-His-Arg-Gly-Ser-Pro-Thr
Mutant 3: Met-Cys-Ile-Val-Val-Val-Gln-His _
Use this information to answer the following questions:
Using the genetic coding dictionary, predict the type of mutation that led to each altered protein.
Shown below are two homologous lengths of the alpha and beta chains of human hemoglobin. Consult a genetic code dictionary (Figure 13.7), and determine how many amino acid substitutions may have occurred as a result of a single nucleotide substitution. For any that cannot occur as a result of a single change, determine the minimal mutational distance.
Alpha: ala val ala his val asp asp met pro
Beta: gly leu ala his leu asp asn leu lys
Mutations in the IL2RG gene cause approximately 30 percent of severe combined immunodeficiency disorder (SCID) cases in humans. These mutations result in alterations to a protein component of cytokine receptors that are essential for proper development of the immune system. The IL2RG gene is composed of eight exons and contains upstream and downstream sequences that are necessary for proper transcription and translation. Below are some of the mutations observed. For each, explain its likely influence on the IL2RG gene product (assume its length to be 375 amino acids).
Nonsense mutation in a coding region
Mutations in the IL2RG gene cause approximately 30 percent of severe combined immunodeficiency disorder (SCID) cases in humans. These mutations result in alterations to a protein component of cytokine receptors that are essential for proper development of the immune system. The IL2RG gene is composed of eight exons and contains upstream and downstream sequences that are necessary for proper transcription and translation. Below are some of the mutations observed. For each, explain its likely influence on the IL2RG gene product (assume its length to be 375 amino acids).
Insertion in Exon 1, causing frameshift
Mutations in the IL2RG gene cause approximately 30 percent of severe combined immunodeficiency disorder (SCID) cases in humans. These mutations result in alterations to a protein component of cytokine receptors that are essential for proper development of the immune system. The IL2RG gene is composed of eight exons and contains upstream and downstream sequences that are necessary for proper transcription and translation. Below are some of the mutations observed. For each, explain its likely influence on the IL2RG gene product (assume its length to be 375 amino acids).
Insertion in Exon 7, causing frameshift
