Here are the essential concepts you must grasp in order to answer the question correctly.
End-Replication Problem
The end-replication problem refers to the difficulty eukaryotic cells face in fully replicating the ends of linear chromosomes during DNA replication. DNA polymerases can only synthesize DNA in one direction and require a primer, which means that the very end of the chromosome cannot be completely copied, leading to progressive shortening of the DNA with each cell division.
Recommended video:
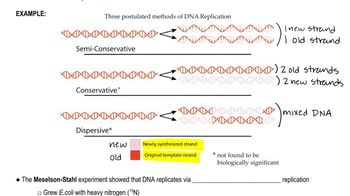
Semiconservative Replication
Telomeres
Telomeres are repetitive nucleotide sequences located at the ends of eukaryotic chromosomes that protect them from degradation and prevent the loss of essential genetic information during replication. They act as a buffer zone, ensuring that the coding regions of the DNA are not affected by the end-replication problem, thus maintaining genomic stability.
Recommended video:
Telomerase
Telomerase is an enzyme that adds repetitive nucleotide sequences to the telomeres, counteracting the end-replication problem. It is particularly active in stem cells and germ cells, allowing these cells to maintain their telomere length and continue dividing without losing vital genetic information, thus playing a crucial role in cellular aging and cancer biology.
Recommended video: