Predict what would happen to regulation of the lac operon if the lacI gene were moved 50,000 nucleotides upstream of its normal location.
In a mutant that lacks adenylyl cyclase, the enzyme that synthesizes cAMP, predict which of the following conditions of extracellular lactose and glucose would cause regulation of the lac operon to differ from that of wild-type cells. a. no lactose, no glucose b. no lactose, abundant glucose c. abundant lactose, no glucose d. abundant lactose, abundant glucose
 Verified step by step guidance
Verified step by step guidance
Verified Solution
Key Concepts
Lac Operon Regulation
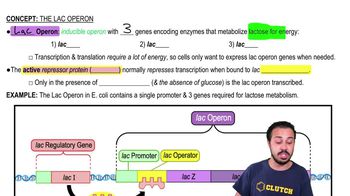
cAMP and Adenylyl Cyclase

Catabolite Repression
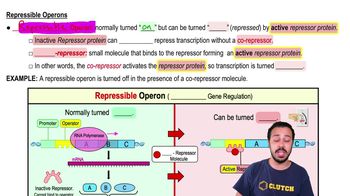
Explain why it makes sense for the lexA regulatory gene of the SOS regulon to be expressed constitutively.
IPTG is a molecule with a structure much like lactose. IPTG can be transported into cells by galactoside permease and can bind to the lac repressor protein. However, unlike lactose, IPTG is not broken down by ββ-galactosidase. Predict what would occur to lac operon regulation if IPTG were added to E. coli growth medium containing no glucose or lactose.
X-gal is a colorless, lactose-like molecule that can be split into two fragments by ββ-galactosidase. One of these product molecules creates a blue color. The photograph here shows E. coli colonies growing in a medium that contains X-gal. Find three colonies whose cells have functioning copies of ββ-galactosidase. Find three colonies whose cells might have mutations in the lacZ or the lacY genes. Suppose you analyze the protein-coding sequence of the lacZ and lacY genes of cells from the three mutant colonies and find that these sequences are wild type (normal). What other region of the lac operon might be altered to account for the mutant phenotype of these colonies?
The light-producing genes of V. fischeri are organized in an operon that is under positive control by an activator protein called LuxR. Would you expect the genes of this operon to be transcribed when LuxR is bound or not bound to a DNA regulatory sequence? Explain.
The diagram shown here is a model of the gene regulatory circuit for light production by V. fischeri cells. The lux operon contains genes for luminescence (luxCDABE) and a gene, luxI, that encodes an enzyme that catalyzes the production of an inducer. This inducer easily moves back and forth across the plasma membrane and acts as a signaling molecule. The lux operon is never completely turned off. The luxR gene codes for the activator LuxR. The inducer can bind to LuxR, and when it does, the LuxR–inducer complex can bind to a regulatory site to activate transcription of the lux operon and inhibit transcription of luxR. Explain how this gene regulatory circuit accounts for bacteria emitting light only when they reach a high cell density.
