Table of contents
- 1. Introduction to Biology2h 40m
- 2. Chemistry3h 40m
- 3. Water1h 26m
- 4. Biomolecules2h 23m
- 5. Cell Components2h 26m
- 6. The Membrane2h 31m
- 7. Energy and Metabolism2h 0m
- 8. Respiration2h 40m
- 9. Photosynthesis2h 49m
- 10. Cell Signaling59m
- 11. Cell Division2h 47m
- 12. Meiosis2h 0m
- 13. Mendelian Genetics4h 44m
- Introduction to Mendel's Experiments7m
- Genotype vs. Phenotype17m
- Punnett Squares13m
- Mendel's Experiments26m
- Mendel's Laws18m
- Monohybrid Crosses19m
- Test Crosses14m
- Dihybrid Crosses20m
- Punnett Square Probability26m
- Incomplete Dominance vs. Codominance20m
- Epistasis7m
- Non-Mendelian Genetics12m
- Pedigrees6m
- Autosomal Inheritance21m
- Sex-Linked Inheritance43m
- X-Inactivation9m
- 14. DNA Synthesis2h 27m
- 15. Gene Expression3h 20m
- 16. Regulation of Expression3h 31m
- Introduction to Regulation of Gene Expression13m
- Prokaryotic Gene Regulation via Operons27m
- The Lac Operon21m
- Glucose's Impact on Lac Operon25m
- The Trp Operon20m
- Review of the Lac Operon & Trp Operon11m
- Introduction to Eukaryotic Gene Regulation9m
- Eukaryotic Chromatin Modifications16m
- Eukaryotic Transcriptional Control22m
- Eukaryotic Post-Transcriptional Regulation28m
- Eukaryotic Post-Translational Regulation13m
- 17. Viruses37m
- 18. Biotechnology2h 58m
- 19. Genomics17m
- 20. Development1h 5m
- 21. Evolution3h 1m
- 22. Evolution of Populations3h 52m
- 23. Speciation1h 37m
- 24. History of Life on Earth2h 6m
- 25. Phylogeny2h 31m
- 26. Prokaryotes4h 59m
- 27. Protists1h 12m
- 28. Plants1h 22m
- 29. Fungi36m
- 30. Overview of Animals34m
- 31. Invertebrates1h 2m
- 32. Vertebrates50m
- 33. Plant Anatomy1h 3m
- 34. Vascular Plant Transport2m
- 35. Soil37m
- 36. Plant Reproduction47m
- 37. Plant Sensation and Response1h 9m
- 38. Animal Form and Function1h 19m
- 39. Digestive System10m
- 40. Circulatory System1h 57m
- 41. Immune System1h 12m
- 42. Osmoregulation and Excretion50m
- 43. Endocrine System4m
- 44. Animal Reproduction2m
- 45. Nervous System55m
- 46. Sensory Systems46m
- 47. Muscle Systems23m
- 48. Ecology3h 11m
- Introduction to Ecology20m
- Biogeography14m
- Earth's Climate Patterns50m
- Introduction to Terrestrial Biomes10m
- Terrestrial Biomes: Near Equator13m
- Terrestrial Biomes: Temperate Regions10m
- Terrestrial Biomes: Northern Regions15m
- Introduction to Aquatic Biomes27m
- Freshwater Aquatic Biomes14m
- Marine Aquatic Biomes13m
- 49. Animal Behavior28m
- 50. Population Ecology3h 41m
- Introduction to Population Ecology28m
- Population Sampling Methods23m
- Life History12m
- Population Demography17m
- Factors Limiting Population Growth14m
- Introduction to Population Growth Models22m
- Linear Population Growth6m
- Exponential Population Growth29m
- Logistic Population Growth32m
- r/K Selection10m
- The Human Population22m
- 51. Community Ecology2h 46m
- Introduction to Community Ecology2m
- Introduction to Community Interactions9m
- Community Interactions: Competition (-/-)38m
- Community Interactions: Exploitation (+/-)23m
- Community Interactions: Mutualism (+/+) & Commensalism (+/0)9m
- Community Structure35m
- Community Dynamics26m
- Geographic Impact on Communities21m
- 52. Ecosystems2h 36m
- 53. Conservation Biology24m
25. Phylogeny
Phylogeny
Problem 2a
Textbook Question
To apply parsimony to constructing a phylogenetic tree, a. choose the tree that assumes all evolutionary changes are equally probable. b. choose the tree in which the branch points are based on as many shared derived characters as possible. c. choose the tree that represents the fewest evolutionary changes, in either DNA sequences or morphology. d. choose the tree with the fewest branch points.
 Verified step by step guidance
Verified step by step guidance1
Understand the concept of parsimony in phylogenetics, which is a method used to infer relationships among species by finding the simplest explanation for observed data, typically minimizing the number of evolutionary changes.
Review each option to determine how it relates to the principle of parsimony: a) assumes all changes are equally probable does not necessarily minimize changes, b) based on shared derived characters focuses on commonality rather than simplicity, c) represents the fewest changes aligns with minimizing evolutionary changes, d) fewest branch points does not directly relate to the number of changes.
Identify that option c) choose the tree that represents the fewest evolutionary changes, in either DNA sequences or morphology, directly addresses the principle of parsimony by minimizing the total number of evolutionary changes.
Recognize that minimizing changes (option c) is more aligned with parsimony than simply minimizing branch points (option d), as fewer branch points do not necessarily equate to fewer evolutionary changes.
Conclude that the best choice to apply parsimony in constructing a phylogenetic tree is option c) choose the tree that represents the fewest evolutionary changes, in either DNA sequences or morphology.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
50sPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Parsimony in Phylogenetics
Parsimony is a principle used in phylogenetics to select the simplest explanation or tree that requires the least number of evolutionary changes. This approach assumes that the most likely evolutionary pathway is the one that minimizes the total number of changes, whether in DNA sequences or morphological traits. By favoring simpler trees, parsimony helps avoid overfitting the data with unnecessary complexity.
Recommended video:
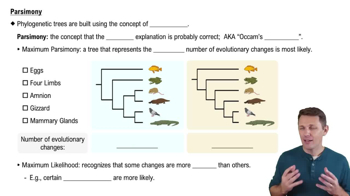
Parsimony
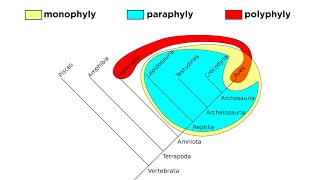
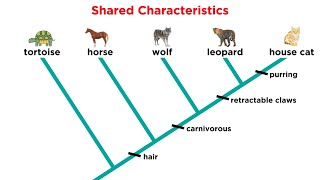
Shared Derived Characters
Shared derived characters, or synapomorphies, are traits that are present in a group of organisms but absent in their distant ancestors. These characters are crucial for constructing phylogenetic trees as they provide evidence of common ancestry and help define evolutionary relationships. The more shared derived characters two species have, the more closely related they are likely to be.
Recommended video:
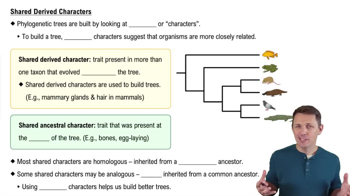
Shared Derived Characters
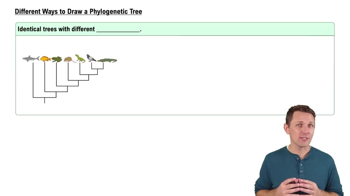
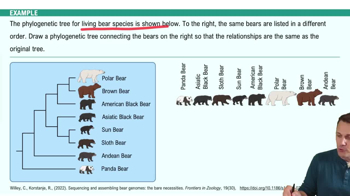
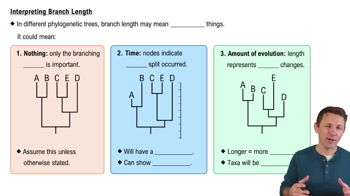
Phylogenetic Tree Construction
Constructing a phylogenetic tree involves organizing species based on their evolutionary relationships, often using data from morphology or genetic sequences. The tree is a visual representation of these relationships, with branch points indicating common ancestors. The goal is to accurately reflect the evolutionary history of the organisms, which can be achieved through methods like parsimony, maximum likelihood, or Bayesian inference.
Recommended video:

Building Phylogenetic Trees Example 2

 7:10m
7:10mWatch next
Master Reading a Phylogenetic Tree with a bite sized video explanation from Bruce Bryan
Start learningRelated Videos
Related Practice