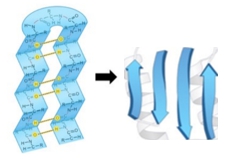
The beta strand is a crucial type of secondary structure in proteins, characterized by an extended zigzag conformation of the protein backbone. This structure is periodic, repeating every two amino acid residues, and is visually represented by the alternating positions of the R groups, which appear to zigzag. The rise, defined as the distance covered per amino acid residue, is approximately 3.5 angstroms for beta strands, significantly greater than the 1.5 angstroms observed in alpha helices. This indicates that beta strands are more extended compared to the coiled structure of alpha helices.
The pitch of a beta strand, which refers to the length of one complete turn of the backbone, measures about 7 angstroms, again longer than the 5.4 angstroms of an alpha helix. To illustrate the differences between these two structures, consider a comparison of five amino acid residues in each conformation. The beta strand, being more extended, has a total length calculated by multiplying the number of amino acids (5) by the rise (3.5 angstroms), resulting in a length of 17.5 angstroms. In contrast, the alpha helix, with the same number of amino acids, yields a length of only 7.5 angstroms when calculated using its rise of 1.5 angstroms.
This comparison highlights the fundamental differences in structure and spatial arrangement between beta strands and alpha helices, emphasizing the extended nature of beta strands versus the coiled configuration of alpha helices. Understanding these distinctions is essential for grasping protein structure and function.