Alright guys. Now I want to talk about the relationship between the equilibrium constant and the pKa. Alright. So remember that in general chemistry we are always talking about something called pH. Okay? And pH was used to measure what? Do you guys remember? It was actually used to measure the concentration of hydronium ions in a solution. Oops. Not is. In solution. Okay. So I know that's really specific, but it was used to basically measure how acidic a solution was. How acidic or how basic. Okay. But it turns out that in organic chemistry, we don't care about the solution at all. We don’t care if it's acidic or if it's basic at all. What we care about is the actual molecule itself and what I care about is how likely is that molecule to donate a proton or how likely is that molecule to accept a proton. pH doesn't tell me that. pH just tells me how many H+ ions are circulating in this test tube. I don't care about that. I care about the molecules inside. I care how much they are likely to give away a proton. Okay? So that means that we're going to use a different measure. We're going to use the negative log of the equilibrium constant or the dissociation constant. Remember that dissociation constant has to do with how likely a bond is to break. Okay? So we're going to use the negative log of how likely that is to happen to figure out the tendency of a molecule to donate protons. Okay? And that's what we actually care about. So that's why in general chemistry you use pH, but now in organic chemistry, we're going to use pKa instead. Okay?
- 1. A Review of General Chemistry5h 5m
- Summary23m
- Intro to Organic Chemistry5m
- Atomic Structure16m
- Wave Function9m
- Molecular Orbitals17m
- Sigma and Pi Bonds9m
- Octet Rule12m
- Bonding Preferences12m
- Formal Charges6m
- Skeletal Structure14m
- Lewis Structure20m
- Condensed Structural Formula15m
- Degrees of Unsaturation15m
- Constitutional Isomers14m
- Resonance Structures46m
- Hybridization23m
- Molecular Geometry16m
- Electronegativity22m
- 2. Molecular Representations1h 14m
- 3. Acids and Bases2h 46m
- 4. Alkanes and Cycloalkanes4h 19m
- IUPAC Naming29m
- Alkyl Groups13m
- Naming Cycloalkanes10m
- Naming Bicyclic Compounds10m
- Naming Alkyl Halides7m
- Naming Alkenes3m
- Naming Alcohols8m
- Naming Amines15m
- Cis vs Trans21m
- Conformational Isomers13m
- Newman Projections14m
- Drawing Newman Projections16m
- Barrier To Rotation7m
- Ring Strain8m
- Axial vs Equatorial7m
- Cis vs Trans Conformations4m
- Equatorial Preference14m
- Chair Flip9m
- Calculating Energy Difference Between Chair Conformations17m
- A-Values17m
- Decalin7m
- 5. Chirality3h 39m
- Constitutional Isomers vs. Stereoisomers9m
- Chirality12m
- Test 1:Plane of Symmetry7m
- Test 2:Stereocenter Test17m
- R and S Configuration43m
- Enantiomers vs. Diastereomers13m
- Atropisomers9m
- Meso Compound12m
- Test 3:Disubstituted Cycloalkanes13m
- What is the Relationship Between Isomers?16m
- Fischer Projection10m
- R and S of Fischer Projections7m
- Optical Activity5m
- Enantiomeric Excess20m
- Calculations with Enantiomeric Percentages11m
- Non-Carbon Chiral Centers8m
- 6. Thermodynamics and Kinetics1h 22m
- 7. Substitution Reactions1h 48m
- 8. Elimination Reactions2h 30m
- 9. Alkenes and Alkynes2h 9m
- 10. Addition Reactions3h 18m
- Addition Reaction6m
- Markovnikov5m
- Hydrohalogenation6m
- Acid-Catalyzed Hydration17m
- Oxymercuration15m
- Hydroboration26m
- Hydrogenation6m
- Halogenation6m
- Halohydrin12m
- Carbene12m
- Epoxidation8m
- Epoxide Reactions9m
- Dihydroxylation8m
- Ozonolysis7m
- Ozonolysis Full Mechanism24m
- Oxidative Cleavage3m
- Alkyne Oxidative Cleavage6m
- Alkyne Hydrohalogenation3m
- Alkyne Halogenation2m
- Alkyne Hydration6m
- Alkyne Hydroboration2m
- 11. Radical Reactions1h 58m
- 12. Alcohols, Ethers, Epoxides and Thiols2h 42m
- Alcohol Nomenclature4m
- Naming Ethers6m
- Naming Epoxides18m
- Naming Thiols11m
- Alcohol Synthesis7m
- Leaving Group Conversions - Using HX11m
- Leaving Group Conversions - SOCl2 and PBr313m
- Leaving Group Conversions - Sulfonyl Chlorides7m
- Leaving Group Conversions Summary4m
- Williamson Ether Synthesis3m
- Making Ethers - Alkoxymercuration4m
- Making Ethers - Alcohol Condensation4m
- Making Ethers - Acid-Catalyzed Alkoxylation4m
- Making Ethers - Cumulative Practice10m
- Ether Cleavage8m
- Alcohol Protecting Groups3m
- t-Butyl Ether Protecting Groups5m
- Silyl Ether Protecting Groups10m
- Sharpless Epoxidation9m
- Thiol Reactions6m
- Sulfide Oxidation4m
- 13. Alcohols and Carbonyl Compounds2h 17m
- 14. Synthetic Techniques1h 26m
- 15. Analytical Techniques:IR, NMR, Mass Spect7h 3m
- Purpose of Analytical Techniques5m
- Infrared Spectroscopy16m
- Infrared Spectroscopy Table31m
- IR Spect:Drawing Spectra40m
- IR Spect:Extra Practice26m
- NMR Spectroscopy10m
- 1H NMR:Number of Signals26m
- 1H NMR:Q-Test26m
- 1H NMR:E/Z Diastereoisomerism8m
- H NMR Table24m
- 1H NMR:Spin-Splitting (N + 1) Rule22m
- 1H NMR:Spin-Splitting Simple Tree Diagrams11m
- 1H NMR:Spin-Splitting Complex Tree Diagrams12m
- 1H NMR:Spin-Splitting Patterns8m
- NMR Integration18m
- NMR Practice14m
- Carbon NMR4m
- Structure Determination without Mass Spect47m
- Mass Spectrometry12m
- Mass Spect:Fragmentation28m
- Mass Spect:Isotopes27m
- 16. Conjugated Systems6h 13m
- Conjugation Chemistry13m
- Stability of Conjugated Intermediates4m
- Allylic Halogenation12m
- Reactions at the Allylic Position39m
- Conjugated Hydrohalogenation (1,2 vs 1,4 addition)26m
- Diels-Alder Reaction9m
- Diels-Alder Forming Bridged Products11m
- Diels-Alder Retrosynthesis8m
- Molecular Orbital Theory9m
- Drawing Atomic Orbitals6m
- Drawing Molecular Orbitals17m
- HOMO LUMO4m
- Orbital Diagram:3-atoms- Allylic Ions13m
- Orbital Diagram:4-atoms- 1,3-butadiene11m
- Orbital Diagram:5-atoms- Allylic Ions10m
- Orbital Diagram:6-atoms- 1,3,5-hexatriene13m
- Orbital Diagram:Excited States4m
- Pericyclic Reaction10m
- Thermal Cycloaddition Reactions26m
- Photochemical Cycloaddition Reactions26m
- Thermal Electrocyclic Reactions14m
- Photochemical Electrocyclic Reactions10m
- Cumulative Electrocyclic Problems25m
- Sigmatropic Rearrangement17m
- Cope Rearrangement9m
- Claisen Rearrangement15m
- 17. Ultraviolet Spectroscopy51m
- 18. Aromaticity2h 34m
- 19. Reactions of Aromatics: EAS and Beyond5h 1m
- Electrophilic Aromatic Substitution9m
- Benzene Reactions11m
- EAS:Halogenation Mechanism6m
- EAS:Nitration Mechanism9m
- EAS:Friedel-Crafts Alkylation Mechanism6m
- EAS:Friedel-Crafts Acylation Mechanism5m
- EAS:Any Carbocation Mechanism7m
- Electron Withdrawing Groups22m
- EAS:Ortho vs. Para Positions4m
- Acylation of Aniline9m
- Limitations of Friedel-Crafts Alkyation19m
- Advantages of Friedel-Crafts Acylation6m
- Blocking Groups - Sulfonic Acid12m
- EAS:Synergistic and Competitive Groups13m
- Side-Chain Halogenation6m
- Side-Chain Oxidation4m
- Reactions at Benzylic Positions31m
- Birch Reduction10m
- EAS:Sequence Groups4m
- EAS:Retrosynthesis29m
- Diazo Replacement Reactions6m
- Diazo Sequence Groups5m
- Diazo Retrosynthesis13m
- Nucleophilic Aromatic Substitution28m
- Benzyne16m
- 20. Phenols55m
- 21. Aldehydes and Ketones: Nucleophilic Addition4h 56m
- Naming Aldehydes8m
- Naming Ketones7m
- Oxidizing and Reducing Agents9m
- Oxidation of Alcohols28m
- Ozonolysis7m
- DIBAL5m
- Alkyne Hydration9m
- Nucleophilic Addition8m
- Cyanohydrin11m
- Organometallics on Ketones19m
- Overview of Nucleophilic Addition of Solvents13m
- Hydrates6m
- Hemiacetal9m
- Acetal12m
- Acetal Protecting Group16m
- Thioacetal6m
- Imine vs Enamine15m
- Addition of Amine Derivatives5m
- Wolff Kishner Reduction7m
- Baeyer-Villiger Oxidation39m
- Acid Chloride to Ketone7m
- Nitrile to Ketone9m
- Wittig Reaction18m
- Ketone and Aldehyde Synthesis Reactions14m
- 22. Carboxylic Acid Derivatives: NAS2h 51m
- Carboxylic Acid Derivatives7m
- Naming Carboxylic Acids9m
- Diacid Nomenclature6m
- Naming Esters5m
- Naming Nitriles3m
- Acid Chloride Nomenclature5m
- Naming Anhydrides7m
- Naming Amides5m
- Nucleophilic Acyl Substitution18m
- Carboxylic Acid to Acid Chloride6m
- Fischer Esterification5m
- Acid-Catalyzed Ester Hydrolysis4m
- Saponification3m
- Transesterification5m
- Lactones, Lactams and Cyclization Reactions10m
- Carboxylation5m
- Decarboxylation Mechanism14m
- Review of Nitriles46m
- 23. The Chemistry of Thioesters, Phophate Ester and Phosphate Anhydrides1h 10m
- 24. Enolate Chemistry: Reactions at the Alpha-Carbon1h 53m
- Tautomerization9m
- Tautomers of Dicarbonyl Compounds6m
- Enolate4m
- Acid-Catalyzed Alpha-Halogentation4m
- Base-Catalyzed Alpha-Halogentation3m
- Haloform Reaction8m
- Hell-Volhard-Zelinski Reaction3m
- Overview of Alpha-Alkylations and Acylations5m
- Enolate Alkylation and Acylation12m
- Enamine Alkylation and Acylation16m
- Beta-Dicarbonyl Synthesis Pathway7m
- Acetoacetic Ester Synthesis13m
- Malonic Ester Synthesis15m
- 25. Condensation Chemistry2h 9m
- 26. Amines1h 43m
- 27. Heterocycles2h 0m
- Nomenclature of Heterocycles15m
- Acid-Base Properties of Nitrogen Heterocycles10m
- Reactions of Pyrrole, Furan, and Thiophene13m
- Directing Effects in Substituted Pyrroles, Furans, and Thiophenes16m
- Addition Reactions of Furan8m
- EAS Reactions of Pyridine17m
- SNAr Reactions of Pyridine18m
- Side-Chain Reactions of Substituted Pyridines20m
- 28. Carbohydrates5h 53m
- Monosaccharide20m
- Monosaccharides - D and L Isomerism9m
- Monosaccharides - Drawing Fischer Projections18m
- Monosaccharides - Common Structures6m
- Monosaccharides - Forming Cyclic Hemiacetals12m
- Monosaccharides - Cyclization18m
- Monosaccharides - Haworth Projections13m
- Mutarotation11m
- Epimerization9m
- Monosaccharides - Aldose-Ketose Rearrangement8m
- Monosaccharides - Alkylation10m
- Monosaccharides - Acylation7m
- Glycoside6m
- Monosaccharides - N-Glycosides18m
- Monosaccharides - Reduction (Alditols)12m
- Monosaccharides - Weak Oxidation (Aldonic Acid)7m
- Reducing Sugars23m
- Monosaccharides - Strong Oxidation (Aldaric Acid)11m
- Monosaccharides - Oxidative Cleavage27m
- Monosaccharides - Osazones10m
- Monosaccharides - Kiliani-Fischer23m
- Monosaccharides - Wohl Degradation12m
- Monosaccharides - Ruff Degradation12m
- Disaccharide30m
- Polysaccharide11m
- 29. Amino Acids3h 20m
- Proteins and Amino Acids19m
- L and D Amino Acids14m
- Polar Amino Acids14m
- Amino Acid Chart18m
- Acid-Base Properties of Amino Acids33m
- Isoelectric Point14m
- Amino Acid Synthesis: HVZ Method12m
- Synthesis of Amino Acids: Acetamidomalonic Ester Synthesis16m
- Synthesis of Amino Acids: N-Phthalimidomalonic Ester Synthesis13m
- Synthesis of Amino Acids: Strecker Synthesis13m
- Reactions of Amino Acids: Esterification7m
- Reactions of Amino Acids: Acylation3m
- Reactions of Amino Acids: Hydrogenolysis6m
- Reactions of Amino Acids: Ninhydrin Test11m
- 30. Peptides and Proteins2h 42m
- Peptides12m
- Primary Protein Structure4m
- Secondary Protein Structure17m
- Tertiary Protein Structure11m
- Disulfide Bonds17m
- Quaternary Protein Structure10m
- Summary of Protein Structure7m
- Intro to Peptide Sequencing2m
- Peptide Sequencing: Partial Hydrolysis25m
- Peptide Sequencing: Partial Hydrolysis with Cyanogen Bromide7m
- Peptide Sequencing: Edman Degradation28m
- Merrifield Solid-Phase Peptide Synthesis18m
- 32. Lipids 2h 50m
- 34. Nucleic Acids1h 32m
- 35. Transition Metals5h 33m
- Electron Configuration of Elements45m
- Coordination Complexes20m
- Ligands24m
- Electron Counting10m
- The 18 and 16 Electron Rule13m
- Cross-Coupling General Reactions40m
- Heck Reaction40m
- Stille Reaction13m
- Suzuki Reaction25m
- Sonogashira Coupling Reaction17m
- Fukuyama Coupling Reaction15m
- Kumada Coupling Reaction13m
- Negishi Coupling Reaction16m
- Buchwald-Hartwig Amination Reaction19m
- Eglinton Reaction17m
- 36. Synthetic Polymers1h 49m
- Introduction to Polymers6m
- Chain-Growth Polymers10m
- Radical Polymerization15m
- Cationic Polymerization8m
- Anionic Polymerization8m
- Polymer Stereochemistry3m
- Ziegler-Natta Polymerization4m
- Copolymers6m
- Step-Growth Polymers11m
- Step-Growth Polymers: Urethane6m
- Step-Growth Polymers: Polyurethane Mechanism10m
- Step-Growth Polymers: Epoxy Resin8m
- Polymers Structure and Properties8m
Equilibrium Constant - Online Tutor, Practice Problems & Exam Prep
 Created using AI
Created using AINow that we understand what an acid is, we need a method of quantifying which acids are stronger and which are weaker. pH doesn’t work for this, let me explain why:
pH vs. pKa
Why we use pKa instead of pH.
Video transcript
The Ka (dissociation constant) describes the tendency of a molecule to break apart. In the case of acids, that specifically means donating protons, which is exactly what we are interested in knowing!
The relationship between equilibrium constant and pKa.
Video transcript
Let's just talk about stuff that you guys should remember, that strong acids are going to have a high dissociation constant. That means that they're very likely to dissociate fully. Okay? And that means, right, they fully dissociate in an aqueous solution. Remember that weak acids are going to have a smaller dissociation constant and what that means is that they're only going to partially dissociate in an aqueous solution. And that makes a huge difference because that means that they're going to have different pKa's. They're going to have different tendencies to donate protons. Now let's remember what is pKa. Well p, remember, stands for the negative log base 10. And then, remember that Ka stands just basically for products over reactants.
Now, I'm not going to make you guys calculate every single thing. But, check it out. I mean, products in this case are just dissociating into H+. And reactants are what happens before it fully dissociates. Does that make sense? So the Ka is basically the ratio of how much of my acid is going to actually become a proton. And that's what I care about. So therefore, if we're taking the negative log of this Ka, what that means is that the higher the Ka, basically the higher the chances of the molecule breaking apart and making ions, the lower the pKa is going to be.
So just like we had in pH, remember that your strongest acid was actually the one that was the lowest pH. It was, like, close to 0. Remember that basically pH is on a scale of 0 to 14 and remember that down here was the very acidic solution and then over at 14 was the very basic solution. So remember that if it was very acidic, it would have a very low pH, And in the same way, pKa is going to do the same thing. So your strongest acids are always going to have the lowest numbers for pKa. Does that make sense? And it's because we're using the negative log, not the positive. So it's always going to be the opposite. Cool so far? Awesome.
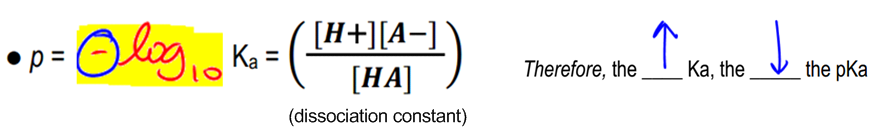
The pH scale vs. the pKa scale.
Video transcript
Now what I want to do is go over the similarity of the pH scale and the pKa scale. So, for the pH scale, like I said, 0 is very acidic, 14 is very basic, and 7 is neutral. Remember that 7 is just water, neutral, right in the middle. For pKa, we have something very similar, but for pKa, it's just going to be a different scale and it's going to mean something different. So, for pKa, about the lowest pKa that you can get without being a crazy molecule is around negative 10. And these are going to be the most amazing acids. Remember that pKa has to do with how much it wants to be an acid. So, negative 10 is going to be like the most amazing acids ever. It turns out that the scale is about from negative 10 to about 50. So, 50 would be about the same thing as me saying, like, a really high number for pH. And what I'm wondering is what do you think 50 means? Do you think that means that it's a very good base? And the answer is no. It has nothing to do with basicity because we're just trying to see how likely it is to dissociate into an acid. So, what is 50? 50 is just going to be very poor acids. They are terrible at dissociating. So, basically, 50 doesn't mean that you're basic; it just means that you're really bad at being an acid. It means that as an acid, you're never going to dissociate. Then, what is 16? 16 is actually water, and water is going to be kind of your neutral pKa, where basically anything above water, we're going to say that those are the good acids, and then anything below water, we're going to say that those are the bad acids. Does that make sense? With 50 being the worst of all, so much so that I said it was very poor. Does that make sense? So, basically, when we're talking about pKas, we're always looking at things that are lower than 16 as our strong or as our good acids.
The pH and pKa scales really are completely different. Pardon my French! pKas are obviously something I’m really passionate about.


Calculating pKa
This is the easiest kind of question you could get. Calculating pKa’s just takes some very simple math.
What is the pKa of acetic acid? Hint:take the negative log of the dissociation constant.

Calculate the pKa of acetic acid
Video transcript
So now what I want you guys to do is calculate the pKa's of the following acids using the dissociation constant. Okay? So, in this case, I'm already giving you the dissociation constant. This is really easy. All you have to do is just take the negative log of it. So I'm just really, like, organic chemistry has very little math in it, but I just want you guys to practice this so you remember the definition of negative log. So, in your calculator, what you should do is just type in and you guys can follow along with me. So maybe pause the video if you need to get your calculator. Okay. You guys all got it. You have your iPhone sideways. Perfect. Awesome.
So what we're gonna do is we're gonna say \( 1.75 \times 10^{-5} \). Okay? And we're just going to take the negative log of that. So I'm gonna basically type in that number and then I'm going to type in log base 10, if it says that. Okay. And then I'm going to type in the negative sign. okay? Remember that you basically have, like, a positive-negative button on your calculator. So you're gonna type in the negative sign on your calculator and what that's gonna give you is a pKa of 4.75. Okay? So is that a good acid? What do you guys think? Yeah, that's actually a pretty good acid because remember I said anything below 16 is pretty good. So it turns out this is actually a carboxylic acid. Remember carboxylic acid? So it turns out carboxylic acids are actually pretty good acids. Duh. Right? That's why they're called carboxylic acids.
What is the pKa of ammonium? Hint:take the negative log of the dissociation constant.

Calculating pKa and comparing acidity
Video transcript
Let's try this next one. Notice that it has a dissociation constant of that, slightly different. Go ahead and take the negative log of that number and see what you get. So now what you should have gotten is you should have gotten the number 9.24 if you round it. Okay? And that's the pKa. So now I want to ask you guys, which of these is the stronger acid? Is acetic acid stronger, or is ammonium stronger? And the answer is it has to be acetic acid. Okay? This is my stronger one because of the fact that it has a lower pKa. Okay? In fact, every time that you drop 1 in pKa, that actually means it's 10 times stronger because it's a log scale. So it turns out that acetic acid is actually like probably like almost like 100000 times better of an acid than ammonium. Isn't that crazy? Like, the numbers get really big really fast when it comes to acidity. So I hope that makes sense. So it's just a quick review. What's most important about this entire page is you guys knowing this trend right here where your lowest pKas are going to be your strongest acids, your highest pKas are going to be your worst acids. Never say that they are bases because pKa has only to do with acids and not with anything else. So let me know if I can clear that up for you in any other ways, but if not, let's keep going.
Do you want more practice?
More setsHere’s what students ask on this topic:
What is the relationship between Ka and pKa in organic chemistry?
The relationship between Ka and pKa is crucial in understanding acid strength. Ka, the acid dissociation constant, measures how completely an acid dissociates in solution. A high Ka indicates a strong acid that dissociates fully, while a low Ka indicates a weak acid that only partially dissociates. pKa is the negative logarithm of Ka, given by the equation:
Thus, a lower pKa corresponds to a higher Ka, indicating a stronger acid. This inverse relationship helps chemists understand and compare the acid strengths of different molecules.
 Created using AI
Created using AIHow does pKa differ from pH in measuring acidity?
pKa and pH are both measures related to acidity, but they serve different purposes. pH measures the concentration of hydronium ions (H3O+) in a solution, indicating how acidic or basic the solution is. It ranges from 0 (very acidic) to 14 (very basic), with 7 being neutral. pKa, on the other hand, measures the tendency of a specific molecule to donate a proton, reflecting its intrinsic acid strength. It is the negative logarithm of the acid dissociation constant (Ka). While pH is about the solution's acidity, pKa is about the molecule's proton-donating ability. Lower pKa values indicate stronger acids, similar to how lower pH values indicate higher acidity.
 Created using AI
Created using AIWhy do strong acids have low pKa values?
Strong acids have low pKa values because pKa is the negative logarithm of the acid dissociation constant (Ka). For strong acids, Ka is high, meaning they dissociate completely in solution, releasing a large number of protons (H+). The equation for pKa is:
Because Ka is large for strong acids, the negative logarithm results in a low pKa value. This low pKa indicates a high tendency to donate protons, which is characteristic of strong acids. Therefore, the lower the pKa, the stronger the acid.
 Created using AI
Created using AIWhat is the pKa scale and how is it used in organic chemistry?
The pKa scale is a numerical representation of acid strength, ranging from about -10 to 50. It is used in organic chemistry to compare the proton-donating abilities of different molecules. The scale is inversely related to acid strength: lower pKa values indicate stronger acids, while higher pKa values indicate weaker acids. For example, a pKa of -10 represents a very strong acid, while a pKa of 50 represents a very weak acid. This scale helps chemists predict reaction outcomes, understand molecular behavior, and design synthesis pathways by providing a clear measure of how likely a molecule is to donate a proton.
 Created using AI
Created using AIHow do you calculate pKa from Ka?
To calculate pKa from Ka, you use the following equation:
First, determine the value of Ka, the acid dissociation constant, which represents the ratio of the concentration of dissociated ions to the concentration of the undissociated acid. Once you have Ka, take the negative logarithm (base 10) of this value to find pKa. For example, if Ka = 1.0 x 10-5, then:
This calculation shows that the pKa is 5, indicating the acid's strength.
 Created using AI
Created using AIYour Organic Chemistry tutors
- 1. CH3CH2OH+NH3⇌CH3CH2O−++NH4 or CH3OH+NH3⇌CH3O−++NH4 b. Which of the four reactions has the most favorable e...
- b. Determine the exact pKa values, using a calculator. c. Which is the strongest acid?
- a. Given the Ka values, estimate the pKa value of each of the following acids without using a calculator (that...
- (••) For the following acid–base pairs, (iii) predict the favored side of equilibrium; (iv) calculate ; (...
- Calculate K_eq for these acid–base reactions. (c)
- Given that the indicated pKₐ values correspond to the acid dissociation reactions shown, calculate the ratio o...
- Hydrogen gas (H₂) has a relatively high pKₐ value. Is it a stable or unstable acid? Do you expect it to partic...
- For the following acid–base reaction, (b) calculate the equilibrium constant.
- (•) For each of the following acid–base reactions, (ii) calculate K_eq , If a pKₐ is not one of the ten c...
- Estimate the K_eq for the following reactions based on the stability of the anions involved. (c)
- Estimate the K_eq for the following reactions based on the stability of the anions involved. (b)
- (••) Using pKₐ values, calculate Kₑq for the following acid–base reaction. ⇌
- Using pKₐ values, calculate the equilibrium constants for the following acid–base reactions. (b)
- Calculate Kₑq for the following acid–base reactions. (b)
- (•) Given the pKb, calculate the pKa of the conjugate acid. c.
- (•) Calculate the equilibrium constant for each of the acid–base reactions shown. b.
- Predict the Kₑq for the following acid–base reaction.
- We usually calculate Kₑq for acid–base reactions using pKₐ values. (a) Derive an equation to calculate Kₑq usi...
- (•) Given the pKb, calculate the pKa of the conjugate acid. b.
- Calculate Keq for these acid–base reactions.(d) <IMAGE>
- (•••) THINKING AHEAD Using pKₐ values for the conjugate acids of the bases on each side of the reaction arrow,...
- Calculate Kₑq for the acid–base reaction shown. Which side is favored and why?<IMAGE>
- Write the K_eq expression for the following acid–base reactions. [You don't need to calculate K_eq here.](c) &...
- (••••) Parts (a)–(d) of this assessment assist in the development of what will become a common theme in organi...
- Calculate the equilibrium constant for the reaction of ethylamine and propanoic acid. Which side is favored?&l...
- How do you know that a proton with a low pKₐ value is acidic (besides 'I just remember')?
- (••) For the following acid–base pairs, (iv) calculate Keq; (e) <IMAGE>
- Given that the indicated pKₐ values correspond to the acid dissociation reactions shown, calculate the ratio o...
- Using pKₐ values, calculate an approximate K_eq value for the following substitution reaction. <IMAGE>
- ••••) THINKING AHEAD Amino acids exist predominantly in one of the following forms. Which is it? Explain your ...
- At what pH is the concentration of a compound, with a pKa=4.6, 10 times greater in its basic form than in its ...
- b. At what pH does 80% of the acid exist in its acidic form?
- The pKa of a protonated alcohol is about -2.5, and the pKa of an alcohol is about 15. Therefore, as long as th...
- The of protonated acetone is about and the of protonated hydroxylamine is 6.0.a. In a reaction with hydroxylam...
- The pKa of acetic acid in water is 4.76. What effect will a decrease in the polarity of the solvent have on th...
- Calculate the equilibrium constant for the acid–base reaction between the reactants in each of the following p...
- Which reaction in Problem 23 has the smallest equilibrium constant?
- Given that pH+pOH=14 and that the concentration of water in a solution of water is 55.5 M, show that the pKa o...
- For each of the following compounds, draw the form that predominates at pH=3, pH=6, pH=10, and pH=14: B. CH3C...
- A naturally occurring amino acid such as alanine has a group that is a carboxylic acid and a group that is a p...
- For each of the following compounds (here shown in their acidic forms), write the form that predominates in a...
- At what pH is the concentration of a compound, with a pKa=8.4, 100 times greater in its acidic form than in it...
- a. If an acid with a pKa of 5.3 is in an aqueous solution of pH 5.7, what percentage of the acid is present in...
- For each of the following compounds (here shown in their acidic forms), write the form that predominates in a...
- (••••) Parts (a)–(d) of this assessment assist in the development of what will become a common theme in organi...
- For each of the following compounds (here shown in their acidic forms), write the form that predominates in a...
- As long as the pH is not less than ___________, at least 50% of a protonated amine with a pKa value of 10.4 wi...
- a. Indicate whether a protonated amine (RN+H3) with a pKa value of 9 has more charged or more neutral molecule...
- For each of the following compounds, indicate the pH at whicha. 50% of the compound is in a form that possesse...
- For each of the following compounds, draw the form that predominates at pH=3, pH=6, pH=10, and pH=14:A. CH3COO...
- For each of the following compounds (here shown in their acidic forms), write the form that predominates in a...
- b. Indicate whether an alcohol (ROH) with a pKa value of 15 has more charged or more neutral molecules in a so...
- b. Indicate whether an alcohol (ROH) with a pKa value of 15 has more charged or more neutral molecules in a so...
- A naturally occurring amino acid such as alanine has a group that is a carboxylic acid and a group that is a p...
- For the following acid–base reaction, (c) calculate the ratio of butan-2-ol to 2-butoxide.
- What percent of imidazole is protonated at physiological pH (7.4)?

