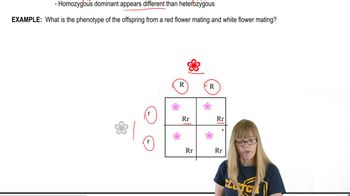
Textbook Question
In rabbits, a series of multiple alleles controls coat color in the following way: C is dominant to all other alleles and causes full color. The chinchilla phenotype is due to the c^ch allele, which is dominant to all alleles other than C. The c^h allele, dominant only to (albino), results in the Himalayan coat color. Thus, the order of dominance is C > c^ch > c^h > c^a. For each of the following three cases, the phenotypes of the P₁ generations of two crosses are shown, as well as the phenotype of one member of the F₁ generation.
For each case, determine the genotypes of the P₁ generation and the F₁ offspring, and predict the results of making each indicated cross between F₁ individuals.
3841
views