In humans, congenital heart disease is a common birth defect that affects approximately 1 out of 125 live births. Using reverse transcription PCR (RT-PCR) Samir Zaidi and colleagues [(2013) Nature 498:220.223] determined that approximately 10 percent of the cases resulted from point mutations, often involving histone function. To capture products of gene expression in developing hearts, they used oligo(dT) in their reverse transcription protocol.
Compared with oligo(dT) primers, a pool of random sequence primers requires a trickier assessment of annealing temperature. Why?
The U.S. Department of Justice has established a database that catalogs PCR amplification products from short tandem repeats of the Y chromosome (Y-STRs) in humans. The database contains polymorphisms of five U.S. ethnic groups (African-Americans, European Americans, Hispanics, Native Americans, and Asian-Americans) as well as the worldwide population.
Given that STRs are repeats of varying lengths, for example (TCTG)₉₋₁₇ or (TAT)₆₋₁₄, explain how PCR could reveal differences (polymorphisms) among individuals. How could the Department of Justice make use of those differences?
 Verified step by step guidance
Verified step by step guidance
Verified Solution
Key Concepts
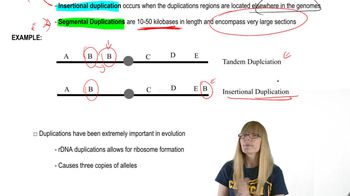
Short Tandem Repeats (STRs)
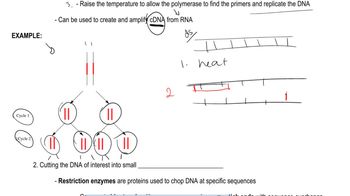
Polymerase Chain Reaction (PCR)
Forensic Applications of Y-STRs

In humans, congenital heart disease is a common birth defect that affects approximately 1 out of 125 live births. Using reverse transcription PCR (RT-PCR) Samir Zaidi and colleagues [(2013) Nature 498:220.223] determined that approximately 10 percent of the cases resulted from point mutations, often involving histone function. To capture products of gene expression in developing hearts, they used oligo(dT) in their reverse transcription protocol.
If one were interested in comparing the quantitative distribution of gene expression in say, the right and left side of a developing heart, how might one proceed using RT-PCR?
The U.S. Department of Justice has established a database that catalogs PCR amplification products from short tandem repeats of the Y chromosome (Y-STRs) in humans. The database contains polymorphisms of five U.S. ethnic groups (African-Americans, European Americans, Hispanics, Native Americans, and Asian-Americans) as well as the worldwide population.
For forensic applications, the probability of a 'match' for a crime scene DNA sample and a suspect's DNA often culminates in a guilty or innocent verdict. How is a 'match' determined, and what are the uses and limitations of such probabilities?
The U.S. Department of Justice has established a database that catalogs PCR amplification products from short tandem repeats of the Y chromosome (Y-STRs) in humans. The database contains polymorphisms of five U.S. ethnic groups (African-Americans, European Americans, Hispanics, Native Americans, and Asian-Americans) as well as the worldwide population.
Y-STRs from the nonrecombining region of the Y chromosome (NRY) have special relevance for forensic purposes. Why?
The U.S. Department of Justice has established a database that catalogs PCR amplification products from short tandem repeats of the Y chromosome (Y-STRs) in humans. The database contains polymorphisms of five U.S. ethnic groups (African-Americans, European Americans, Hispanics, Native Americans, and Asian-Americans) as well as the worldwide population.
What would be the value of knowing the ethnic population differences for Y-STR polymorphisms?
There are a variety of circumstances under which rapid results using multiple markers in PCR amplifications are highly desired, such as in forensics, pathogen analysis, or detection of genetically modified organisms. In multiplex PCR, multiple sets of primers are used, often with less success than when applied to PCR as individual sets. Numerous studies have been conducted to optimize procedures, but each has described the process as time consuming and often unsuccessful. Considering the information given in Problem 30, why should multiplex PCR be any different than single primer set PCR in terms of dependability and ease of optimization?
