Contrast the structure of SINE and LINE DNA sequences. Why are LINEs referred to as retrotransposons?
 Verified step by step guidance
Verified step by step guidance
Verified video answer for a similar problem:
Key Concepts
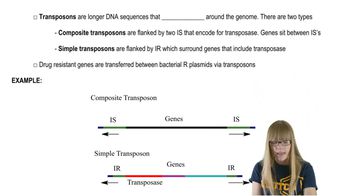
SINE and LINE Sequences

Retrotransposons

Transposition Mechanisms
A significant number of mutations in the HBB gene that cause human β-thalassemia occur within introns or in upstream noncoding sequences. Explain why mutations in these regions often lead to severe disease, although they may not directly alter the coding regions of the gene.
Dominant mutations can be categorized according to whether they increase or decrease the overall activity of a gene or gene product. Although a loss-of-function mutation (a mutation that inactivates the gene product) is usually recessive, for some genes, one dose of the normal gene product, encoded by the normal allele, is not sufficient to produce a normal phenotype. In this case, a loss-of-function mutation in the gene will be dominant, and the gene is said to be haploinsufficient. A second category of dominant mutation is the gain-of-function mutation, which results in a new activity or increased activity or expression of a gene or gene product. The gene therapy technique currently being used in clinical trials involves the 'addition' to somatic cells of a normal copy of a gene. In other words, a normal copy of the gene is inserted into the genome of the mutant somatic cell, but the mutated copy of the gene is not removed or replaced. Will this strategy work for either of the two aforementioned types of dominant mutations?
In 2013 the actress Angelina Jolie elected to have prophylactic double-mastectomy surgery to prevent breast cancer based on a positive test for mutation of the BRCA1 gene. What are some potential positive and negative consequences of this high-profile example of acting on the results of a genetic test?
