This chapter focuses on the mapping of genes on chromosomes, primarily through the use of recombination frequencies. Recombination frequency is defined as the proportion of recombinant offspring produced from a genetic cross, which reflects the likelihood of crossover events between genes during meiosis. The formula to calculate recombination frequency (RF) or map distance is given by:
RF = \(\frac{\text{Number of recombinant offspring}}{\text{Total number of offspring}} \times 100\)
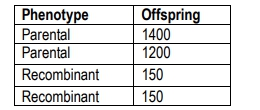
In this context, recombinant offspring are those that exhibit a combination of traits different from their parents, while parental types resemble the original parents. For example, in a cross between a homozygous dominant and a homozygous recessive organism, the resulting F1 generation is heterozygous for both traits. When this F1 generation is crossed with a tester organism, the offspring can be categorized into parental and recombinant types based on their phenotypes.
To illustrate, if the number of recombinant offspring is 154 and the total number of offspring is 2839, the recombination frequency would be calculated as:
RF = \(\frac{154}{2839} \times 100 \approx 10.7\%\)
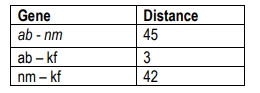
This percentage indicates that the two genes are 10.7 map units apart on the chromosome. A crucial concept to remember is that the closer two genes are on a chromosome, the less likely they are to undergo recombination. This can be visualized with an analogy: if two friends are sitting close together in a hallway, it is unlikely that a stranger will sit between them, whereas if they are far apart, the likelihood of a stranger sitting in between increases.
Understanding the implications of recombination frequency is essential for determining gene linkage. Genes are considered linked if their recombination frequency is less than 50%. Conversely, if the frequency is equal to or greater than 50%, the genes are likely assorting independently, indicating they are on different chromosomes. This independent assortment results in a 50% parental and 50% recombinant distribution of alleles, as no combination can exceed this ratio without violating the principles of Mendelian genetics.
In summary, the key takeaways are that recombination frequency serves as a measure of genetic linkage, with values less than 50% indicating linkage and values at or above 50% suggesting independent assortment. This foundational understanding is critical for genetic mapping and the study of inheritance patterns.