- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
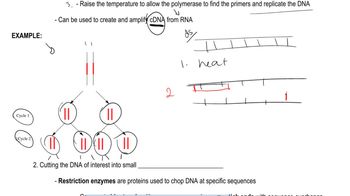
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
To further analyze the CRABS CLAW gene (see Problems 19 and 20), you create a map of the genomic clone. The 11-kb EcoRI fragment is ligated into the EcoRI site of the MCS of the vector shown in Problem 18. You digest the double-stranded form of the genome with several restriction enzymes and obtain the following results. Draw, as far as possible, a map of the genomic clone of CRABS CLAW.
EcoRI 11.0, 3.0 XbaI 4.5, 9.5
EcoRI + XbaI 4.5, 6.5, 3.0 XhoI 13.2, 0.8
EcoRI + XhoI 10.2, 3.0, 0.8 SalI 6.0, 8.0
EcoRI + SalI 6.0, 5.0, 3.0 HindIII 12.0, 1.5, 0.5
EcoRI + HindIII 9.0, 3.0, 1.5, 0.5
What restriction digest would help resolve any ambiguity in the map?
 Verified Solution
Verified Solution