In this video, we're going to begin our lesson on a technique that's called southern blotting, but before we talk about the actual southern blotting technique, it's actually first helpful to talk about radioactive probes. And so in this video, we're going to focus on radioactive probes, and in our next lesson video, we'll talk more about the southern blotting technique. And so what's important to know is that after cloning a gene, the DNA that was cloned can actually be used as a probe to detect the same sequence in an unknown DNA sample. A probe or probes are really just radioactively labeled molecules that are visualized using radioactive detection. And so DNA probes are going to be single-stranded DNA molecules that are going to be complementary to a specific sequence of interest. And so the DNA probe, which is going to be single-stranded and radioactive, when it is bound to the specific sequence of interest, we can detect the presence of a specific sequence of interest by detecting the presence of the radioactive probe. And so if we take a look at our image down below, we can get a better understanding of these radioactive probes. And so here in our example, it's saying radioactive probes are used to identify DNA that is complementary to it in different samples. And so notice in this image over here on the left side, we have 2 different test tubes with different samples. We have sample A over in the left test tube and sample B over here in the right test tube, and both of these samples contain DNA. And so suppose that we want to identify which of these samples has a specific DNA sequence of interest. What we can do is we can use a radioactive DNA probe, which is going to be a single-stranded DNA molecule as you see here, which is going to be radioactively labeled. And so we can take this radioactive DNA probe and we can put it into both of these samples. And under the right conditions, when we incubate the samples with the probe, what will happen is the sample where we can actually detect radioactivity is going to have the sequence of interest, whereas the sample where we do not detect radioactivity will not have the sequence of interest. And here in this image, the radioactivity is represented by the yellow color that you see. And so in terms of having the sequence of interest, because we see radioactivity in sample A, we would say that sample A does have the sequence of interest. So we would say yes here. But sample B, because we do not detect any radioactivity, would mean that it does not have the sequence of interest. And so basically, by using these radioactive probes, we can detect the presence of a specific sequence. And so, we'll be able to see how radioactive probes are going to be important in the technique Southern blotting in our next video. But for now, this here concludes our introduction to radioactive probes and we'll be able to get practice applying these concepts as we move forward. So I'll see you all in our next video.
- 1. Introduction to Biology2h 40m
- 2. Chemistry3h 40m
- 3. Water1h 26m
- 4. Biomolecules2h 23m
- 5. Cell Components2h 26m
- 6. The Membrane2h 31m
- 7. Energy and Metabolism2h 0m
- 8. Respiration2h 40m
- 9. Photosynthesis2h 49m
- 10. Cell Signaling59m
- 11. Cell Division2h 47m
- 12. Meiosis2h 0m
- 13. Mendelian Genetics4h 41m
- Introduction to Mendel's Experiments7m
- Genotype vs. Phenotype17m
- Punnett Squares13m
- Mendel's Experiments26m
- Mendel's Laws18m
- Monohybrid Crosses16m
- Test Crosses14m
- Dihybrid Crosses20m
- Punnett Square Probability26m
- Incomplete Dominance vs. Codominance20m
- Epistasis7m
- Non-Mendelian Genetics12m
- Pedigrees6m
- Autosomal Inheritance21m
- Sex-Linked Inheritance43m
- X-Inactivation9m
- 14. DNA Synthesis2h 27m
- 15. Gene Expression3h 20m
- 16. Regulation of Expression3h 31m
- Introduction to Regulation of Gene Expression13m
- Prokaryotic Gene Regulation via Operons27m
- The Lac Operon21m
- Glucose's Impact on Lac Operon25m
- The Trp Operon20m
- Review of the Lac Operon & Trp Operon11m
- Introduction to Eukaryotic Gene Regulation9m
- Eukaryotic Chromatin Modifications16m
- Eukaryotic Transcriptional Control22m
- Eukaryotic Post-Transcriptional Regulation28m
- Eukaryotic Post-Translational Regulation13m
- 17. Viruses37m
- 18. Biotechnology2h 58m
- 19. Genomics17m
- 20. Development1h 5m
- 21. Evolution3h 1m
- 22. Evolution of Populations3h 52m
- 23. Speciation1h 37m
- 24. History of Life on Earth2h 6m
- 25. Phylogeny40m
- 26. Prokaryotes4h 59m
- 27. Protists1h 6m
- 28. Plants1h 22m
- 29. Fungi36m
- 30. Overview of Animals34m
- 31. Invertebrates1h 2m
- 32. Vertebrates50m
- 33. Plant Anatomy1h 3m
- 34. Vascular Plant Transport2m
- 35. Soil37m
- 36. Plant Reproduction47m
- 37. Plant Sensation and Response1h 9m
- 38. Animal Form and Function1h 19m
- 39. Digestive System10m
- 40. Circulatory System1h 57m
- 41. Immune System1h 12m
- 42. Osmoregulation and Excretion50m
- 43. Endocrine System4m
- 44. Animal Reproduction2m
- 45. Nervous System55m
- 46. Sensory Systems46m
- 47. Muscle Systems23m
- 48. Ecology3h 11m
- Introduction to Ecology20m
- Biogeography14m
- Earth's Climate Patterns50m
- Introduction to Terrestrial Biomes10m
- Terrestrial Biomes: Near Equator13m
- Terrestrial Biomes: Temperate Regions10m
- Terrestrial Biomes: Northern Regions15m
- Introduction to Aquatic Biomes27m
- Freshwater Aquatic Biomes14m
- Marine Aquatic Biomes13m
- 49. Animal Behavior28m
- 50. Population Ecology3h 41m
- Introduction to Population Ecology28m
- Population Sampling Methods23m
- Life History12m
- Population Demography17m
- Factors Limiting Population Growth14m
- Introduction to Population Growth Models22m
- Linear Population Growth6m
- Exponential Population Growth29m
- Logistic Population Growth32m
- r/K Selection10m
- The Human Population22m
- 51. Community Ecology2h 46m
- Introduction to Community Ecology2m
- Introduction to Community Interactions9m
- Community Interactions: Competition (-/-)38m
- Community Interactions: Exploitation (+/-)23m
- Community Interactions: Mutualism (+/+) & Commensalism (+/0)9m
- Community Structure35m
- Community Dynamics26m
- Geographic Impact on Communities21m
- 52. Ecosystems2h 36m
- 53. Conservation Biology24m
Southern Blotting - Online Tutor, Practice Problems & Exam Prep
 Created using AI
Created using AISouthern blotting is a technique used to detect specific DNA sequences using radioactive probes. The process involves fragmenting unknown DNA samples, separating them by size through gel electrophoresis, and denaturing the DNA to single-stranded form. The single-stranded DNA is then transferred to a nitrocellulose filter paper, where it is incubated with radioactive probes that bind to complementary sequences. The presence of these sequences is visualized as bands on the filter paper, indicating the specific DNA of interest. This method is crucial for genetic analysis and research.
Southern Blotting
Video transcript
A southern blotting technique is used to detect a specific ______ sequence from a blood or tissue sample.
A _____ is a single-stranded DNA molecule used in hybridization reactions to detect the presence of a particular gene in an assortment of DNA fragments.
(Hybridization Reactions: reactions that combine two complementary single-stranded DNA molecules)
Steps of Southern Blotting
Video transcript
In this video, we're going to talk about the southern blotting technique. And so southern blotting, again, is a specific technique that's going to be used to rapidly detect a specific DNA sequence by using DNA probes. Now there are other types of blotting that exist. For example, northern blotting, which is going to be used to detect RNA sequences, and Western blotting, which is going to be used to detect specific proteins. But again, in this video, we're going to be mainly focusing on southern blotting, which is used to detect DNA sequences. And so down below, you can see that we're showing you the 5 different steps of southern blotting that are numbered 1 through 5. And of course, these numbers that you see here in the text, 1 through 5, correspond with the numbers that you see down below in the image 1 through 5. And so here in the first step of Southern blotting, we're going to take some unknown DNA samples and fragment them, and then separate those DNA fragments within the unknown DNA sample by size by using gel electrophoresis, which we covered in previous lesson videos. And so if you take a look at our image down below at step number 1, what you can see here is that gel electrophoresis is going to be used to separate fragments of unknown DNA samples. And so you can see our gel electrophoresis down below here. And so in step number 2, what we're going to do is take the DNA on the gel and denature the DNA on the gel so that the DNA on the gel becomes single-stranded DNA or ssDNA, for short. And the way that the DNA is going to be denatured to single-stranded DNA is by incubating the gel with a denaturing buffer. And so if we take a look at our image down below, we can get a better understanding of this. And so what happens is this gel from step number 1 is going to be placed into a container that has multiple different pieces in it. First, this container is going to have the denaturing buffer that's going to help denature the DNA and create single-stranded DNA. But there's also a sponge that's going to be on here. The gel is going to be placed on top of the sponge. Then you'll see a nitrocellulose filter paper which is here in a greenish color, is going to be placed on top of the gel. And then on top of the nitrocellulose filter paper, we have the paper towels that are going to be placed on the very top. And so basically what happens is, the denaturing buffer is going to be absorbed upwards, through all of these different components. And so the denaturing buffer is a liquid that's going to be absorbed into the sponge, which is going to migrate and be absorbed into the gel, which will migrate and be absorbed into the nitrocellulose filter paper, which will then continue to migrate into the paper towel. So basically, the buffer, the denaturing buffer is going to be migrating upwards as you see here in this image. Now the denaturing buffer, the way that it denatures the DNA and creates single-stranded DNA is because the pH of the buffer is actually going to increase, the pH. So the pH is going to be increased, and that causes the DNA to be denatured to single-stranded DNA. Now the filter paper is going to be used to blot the gel, and the blotting here is really just referring to transferring the DNA from the gel to the filter paper. And so it says here that the filter paper will be used to blot the gel and absorb the denaturing buffer. And the denaturing buffer is going to migrate all the way through all of those substances to get to the paper towel stack at the very top. Now, again, what this is going to do is transfer the DNA from the gel to the nitrocellulose filter paper. And the DNA again is going to be denatured as single-stranded DNA. So if we take a look at step number 3, you can see that the single-stranded DNA is transferred over to the nitrocellulose filter paper as the denaturing buffer is being absorbed and migrating through to the paper towels. And so, ultimately what happens is, you can see that the single-stranded DNA fragments are now found on this nitrocellulose filter paper, which we can grab the nitrocellulose filter paper and transfer it to another container here. And the gel, notice, no longer has any more DNA in it because all of the DNA has been denatured to single-stranded DNA and transferred over to the filter paper. Then in step number 4, the filter paper is going to be removed and incubated with radioactive probes that are complementary to the specific sequence of interest that we want to try to detect. And so if we take a look at step number 4, what you can see is we take the nitrocellulose filter paper, and we can put it into a container. Here, we're just showing you a little ziplock baggy, and expose the nitrocellulose filter paper to these DNA probes. And so the nitrocellulose filter paper is going to be incubated with these DNA probes. And recall that the DNA probes are going to be radioactive and they're going to be complementary to the sequence of interest. And so they will bind only to specific fragments of DNA that, it is complementary to. And so in step number 5, the final step here, the radioactive filter paper can be analyzed and visible bands on that filter paper are going to be the ones that bind to the DNA probe in step number 4. And so if you take a look at step number 5 down below, you can see that the DNA is going to be complementary to is the DNA complementary to the probe is visualized. And so what you'll notice is that here we only have a specific set of bands, right here and right here. These bands are the ones that are complementary to the probe. So notice over here on the left-hand side, there are a lot more DNA bands in the filter paper. But the only ones that will actually be visualized are going to be the ones that are complementary to the probe. And so these DNA bands here that are in yellow, these are the only DNA bands of all of the DNA bands that were over here that are actually complementary to the probe. And so these are the bands that are going to have the sequence of interest. And so we can say that lanes number if we were to number these lanes, 1, 2, 3, and 4, lanes 2 and 4, right here and right here are going to be samples that actually contain the sequence of interest that the scientist is interested in. And so this is basically how southern blotting works. It's used to detect specific DNA sequences that are complementary to DNA probes. And so this here concludes our brief introduction to southern blotting, and we'll be able to get some practice applying these concepts as we move forward in our course. So I'll see you all in our next video.
A geneticist wants to see if her patient has Gene X. The geneticist takes a blood sample from her patient and prepares a southern blot. How will the geneticist know if her patient possesses Gene X?
Place the following steps of Southern Blotting in the correct order.
a) ______: Filter paper is incubated with the labeled DNA probe which anneals to the ssDNA fragments.
b) ______: Analyze gel to determine the presence of the DNA sequence of interest.
c) ______: Separate DNA fragments by size using gel electrophoresis.
d) ______: Denature DNA by soaking gel in a basic solution.
e) ______: Fragment unknown DNA sample(s) using restriction enzymes.
Problem Transcript
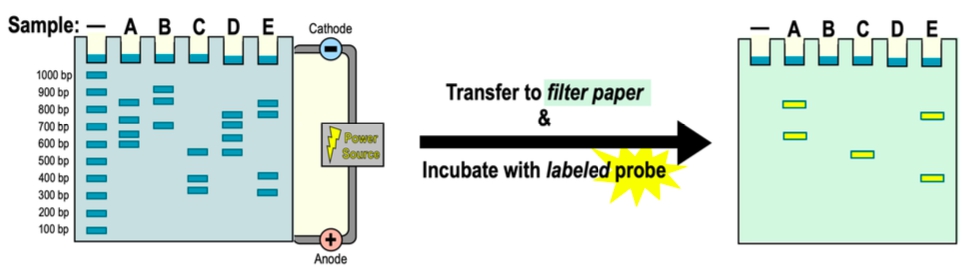
By analyzing the Southern Blot results below, which of the samples contains the gene of interest?
Do you want more practice?
More setsGo over this topic definitions with flashcards
More setsHere’s what students ask on this topic:
What is Southern blotting and how does it work?
Southern blotting is a molecular biology technique used to detect specific DNA sequences in a sample. The process involves several steps: First, DNA is extracted from the sample and fragmented using restriction enzymes. These fragments are then separated by size using gel electrophoresis. Next, the DNA in the gel is denatured to single strands and transferred to a nitrocellulose filter paper. The filter paper is then incubated with radioactive DNA probes that are complementary to the target sequence. If the target sequence is present, the probes will bind to it. The presence of the target DNA is visualized as bands on the filter paper, indicating the specific DNA of interest.
 Created using AI
Created using AIWhat are the steps involved in Southern blotting?
Southern blotting involves five main steps: 1) Fragmentation: DNA samples are fragmented using restriction enzymes. 2) Gel Electrophoresis: The DNA fragments are separated by size using gel electrophoresis. 3) Denaturation: The DNA in the gel is denatured to single strands using a denaturing buffer. 4) Transfer: The single-stranded DNA is transferred to a nitrocellulose filter paper. 5) Hybridization: The filter paper is incubated with radioactive DNA probes that bind to complementary sequences. The presence of the target DNA is visualized as bands on the filter paper.
 Created using AI
Created using AIWhat is the role of radioactive probes in Southern blotting?
In Southern blotting, radioactive probes play a crucial role in detecting specific DNA sequences. These probes are single-stranded DNA molecules that are complementary to the target sequence of interest. They are labeled with a radioactive isotope, allowing for visualization. During the hybridization step, the filter paper containing the single-stranded DNA is incubated with these radioactive probes. If the target sequence is present, the probes will bind to it. The bound probes can then be detected using radioactive detection methods, revealing the presence and location of the target DNA sequence on the filter paper.
 Created using AI
Created using AIHow is gel electrophoresis used in Southern blotting?
Gel electrophoresis is a key step in Southern blotting used to separate DNA fragments by size. After DNA samples are fragmented using restriction enzymes, they are loaded into a gel matrix. An electric current is applied, causing the negatively charged DNA fragments to migrate towards the positive electrode. Smaller fragments move faster and travel further than larger ones, resulting in separation by size. This separation allows for the identification of specific DNA fragments when they are later transferred to a nitrocellulose filter paper and hybridized with radioactive probes.
 Created using AI
Created using AIWhat are the applications of Southern blotting?
Southern blotting has several important applications in molecular biology and genetics. It is used to detect specific DNA sequences in a sample, which is valuable for gene mapping, cloning, and genetic fingerprinting. It can also be used to diagnose genetic disorders by identifying mutations or deletions in genes. Additionally, Southern blotting is employed in forensic science for DNA profiling and in evolutionary biology to study genetic relationships among species. Overall, it is a versatile technique for analyzing DNA and understanding genetic information.
 Created using AI
Created using AI