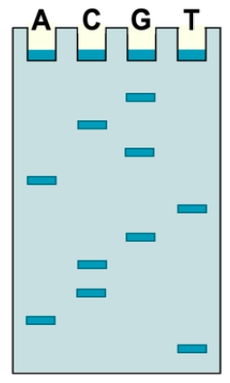
Dideoxy sequencing, also known as Sanger sequencing, is a method used to determine the nucleotide sequence of DNA. To perform this technique, five essential components are required, each playing a crucial role in the sequencing process.
The first component is the unknown template DNA. This is the DNA whose sequence is to be determined, and it serves as the basis for the sequencing reaction. The template DNA is critical because it provides the sequence that will be analyzed.
The second component is DNA polymerase, an enzyme responsible for synthesizing new DNA strands by adding nucleotides to a growing chain. This enzyme is vital for the replication of DNA and is a key player in the dideoxy sequencing process.
Next, DNA primers are needed. These short sequences of nucleotides anneal to the template DNA and provide a starting point for DNA synthesis. In dideoxy sequencing, two specific DNA primers are typically used, similar to their role in the polymerase chain reaction (PCR).
The fourth component consists of the four standard deoxyribonucleotides (dNTPs): deoxyadenosine triphosphate (dATP), deoxycytidine triphosphate (dCTP), deoxyguanosine triphosphate (dGTP), and deoxythymidine triphosphate (dTTP). These nucleotides are the building blocks of DNA and are essential for the synthesis of new DNA strands during the sequencing process.
Finally, the fifth component is a small amount of a single dideoxyribonucleotide (ddNTP). These special nucleotides, such as ddATP, ddCTP, ddGTP, and ddTTP, are crucial for terminating DNA synthesis. The presence of a 3' hydrogen atom in dideoxynucleotides prevents further elongation of the DNA strand, allowing for the generation of fragments of varying lengths that can be analyzed to determine the sequence of the template DNA.
In summary, the five key components required for dideoxy sequencing are unknown template DNA, DNA polymerase, DNA primers, deoxyribonucleotides, and dideoxyribonucleotides. Understanding these components is essential for grasping the mechanics of the dideoxy sequencing process, which will be explored in further detail in subsequent lessons.