Hey guys, in this video, I'm going to show you how to draw molecular orbital diagrams for 3-atom conjugated systems. Let's get started. So guys, because a 3-atom conjugated system is an odd number of atoms, what we're usually going to end up dealing with is an allylic position. Remember that the allyl position or an allylic position means it's the position next to a double bond, and that's usually the case when you have an odd number of atoms because many times there's a double bond and then there's something next to the double bond. That's like the 3rd atom. Okay? Now what we know about allylic positions from previous lessons is that allylic positions are able to resonate. Remember that the allylic position can resonate from one side of the double bond to the other. Okay? So it turns out that the whole idea of resonating an allylic position can actually be explained through molecular orbitals. You already know how to explain it using resonance. You should be able to draw resonance arrows and be able to show how the allylic position can move from one side to the next. But it actually turns out that the fundamental explanation of an allylic position reaction can actually be explained through molecular orbital theory. So that's what we're going to do right here. I'm going to show you guys how molecular orbitals explain the reactive positions of an allylic ion. So here we go. It says simplified LCAO model of a propanyl ion and here what I've drawn is a general propanyl ion. Basically we've got our 3-atom conjugated system. So we have 3 atomic orbitals, like let's just say a, b, and c, right. And what we see is that we have a double bond that we know is going to contribute 1 pi electron for each atom, so we have 12. And then we have this unknown ion or this unknown non-bonding orbital in position C and that I've labeled with a question mark. And the reason is because what I'm about to explain to you applies to any ion, no matter what the identity of that question mark is. Whether it's empty, whether it's an empty orbital or whether it's a radical or whether it's a lone pair. It doesn't matter. What I'm able to teach you applies in all those situations and what the molecular orbital theory would say about this molecule is that how would you structure the molecular orbitals? Well, it says that you would have, first of all, a molecular orbital with 0 nodes, 1 node and 2 nodes, and that we would put those pi electrons in order of Aufbau principle. So we would put the 2 electrons from that first double bond into psi 1, right, which is the most stable the bonding orbital. And then what we would do is we would put whatever is left over, whatever is here, whether it's 0 electrons, 1 electron, or 2 electrons, they would go into psi 2. Does that make sense so far? No matter what the question mark is, we know it's going into psi 2. Now what is unique about psi 2? Look at psi 2. Notice that psi 2 actually has a node at atom B. If this is atom A, atom B and atom C like we had before, we had a, b, and c, right? Notice that there's a node at atom B. What does that mean? What that means is that regardless of the identity of that ion, it cannot react at position B. It can only react at either position A or at position C, but because no electrons ever pass through atom B, you will never find the allylic position reacting there. So what this does is it explains the theory behind the resonance structures that we've learned how to draw for so long. Remember that an allylic position ion can resonate to the other side of the bond, but it can never resonate to the middle. You've never seen that, for example, this, let's say this is a positive charge, you can't move it to the middle. You can only switch places with the double bond, but you can't move it to the middle. And the reason is that the molecular orbital theory states that there's no way that that ion can react at the middle carbon because that orbital doesn't even have any electrons. It can only react at the orbitals with electrons, which would either be orbital C in this case or orbital A in this case, but never B. Isn't that so cool? So what we're going to do next is an example.
- 1. A Review of General Chemistry5h 5m
- Summary23m
- Intro to Organic Chemistry5m
- Atomic Structure16m
- Wave Function9m
- Molecular Orbitals17m
- Sigma and Pi Bonds9m
- Octet Rule12m
- Bonding Preferences12m
- Formal Charges6m
- Skeletal Structure14m
- Lewis Structure20m
- Condensed Structural Formula15m
- Degrees of Unsaturation15m
- Constitutional Isomers14m
- Resonance Structures46m
- Hybridization23m
- Molecular Geometry16m
- Electronegativity22m
- 2. Molecular Representations1h 14m
- 3. Acids and Bases2h 46m
- 4. Alkanes and Cycloalkanes4h 19m
- IUPAC Naming29m
- Alkyl Groups13m
- Naming Cycloalkanes10m
- Naming Bicyclic Compounds10m
- Naming Alkyl Halides7m
- Naming Alkenes3m
- Naming Alcohols8m
- Naming Amines15m
- Cis vs Trans21m
- Conformational Isomers13m
- Newman Projections14m
- Drawing Newman Projections16m
- Barrier To Rotation7m
- Ring Strain8m
- Axial vs Equatorial7m
- Cis vs Trans Conformations4m
- Equatorial Preference14m
- Chair Flip9m
- Calculating Energy Difference Between Chair Conformations17m
- A-Values17m
- Decalin7m
- 5. Chirality3h 39m
- Constitutional Isomers vs. Stereoisomers9m
- Chirality12m
- Test 1:Plane of Symmetry7m
- Test 2:Stereocenter Test17m
- R and S Configuration43m
- Enantiomers vs. Diastereomers13m
- Atropisomers9m
- Meso Compound12m
- Test 3:Disubstituted Cycloalkanes13m
- What is the Relationship Between Isomers?16m
- Fischer Projection10m
- R and S of Fischer Projections7m
- Optical Activity5m
- Enantiomeric Excess20m
- Calculations with Enantiomeric Percentages11m
- Non-Carbon Chiral Centers8m
- 6. Thermodynamics and Kinetics1h 22m
- 7. Substitution Reactions1h 48m
- 8. Elimination Reactions2h 30m
- 9. Alkenes and Alkynes2h 9m
- 10. Addition Reactions3h 18m
- Addition Reaction6m
- Markovnikov5m
- Hydrohalogenation6m
- Acid-Catalyzed Hydration17m
- Oxymercuration15m
- Hydroboration26m
- Hydrogenation6m
- Halogenation6m
- Halohydrin12m
- Carbene12m
- Epoxidation8m
- Epoxide Reactions9m
- Dihydroxylation8m
- Ozonolysis7m
- Ozonolysis Full Mechanism24m
- Oxidative Cleavage3m
- Alkyne Oxidative Cleavage6m
- Alkyne Hydrohalogenation3m
- Alkyne Halogenation2m
- Alkyne Hydration6m
- Alkyne Hydroboration2m
- 11. Radical Reactions1h 58m
- 12. Alcohols, Ethers, Epoxides and Thiols2h 42m
- Alcohol Nomenclature4m
- Naming Ethers6m
- Naming Epoxides18m
- Naming Thiols11m
- Alcohol Synthesis7m
- Leaving Group Conversions - Using HX11m
- Leaving Group Conversions - SOCl2 and PBr313m
- Leaving Group Conversions - Sulfonyl Chlorides7m
- Leaving Group Conversions Summary4m
- Williamson Ether Synthesis3m
- Making Ethers - Alkoxymercuration4m
- Making Ethers - Alcohol Condensation4m
- Making Ethers - Acid-Catalyzed Alkoxylation4m
- Making Ethers - Cumulative Practice10m
- Ether Cleavage8m
- Alcohol Protecting Groups3m
- t-Butyl Ether Protecting Groups5m
- Silyl Ether Protecting Groups10m
- Sharpless Epoxidation9m
- Thiol Reactions6m
- Sulfide Oxidation4m
- 13. Alcohols and Carbonyl Compounds2h 17m
- 14. Synthetic Techniques1h 26m
- 15. Analytical Techniques:IR, NMR, Mass Spect7h 3m
- Purpose of Analytical Techniques5m
- Infrared Spectroscopy16m
- Infrared Spectroscopy Table31m
- IR Spect:Drawing Spectra40m
- IR Spect:Extra Practice26m
- NMR Spectroscopy10m
- 1H NMR:Number of Signals26m
- 1H NMR:Q-Test26m
- 1H NMR:E/Z Diastereoisomerism8m
- H NMR Table24m
- 1H NMR:Spin-Splitting (N + 1) Rule22m
- 1H NMR:Spin-Splitting Simple Tree Diagrams11m
- 1H NMR:Spin-Splitting Complex Tree Diagrams12m
- 1H NMR:Spin-Splitting Patterns8m
- NMR Integration18m
- NMR Practice14m
- Carbon NMR4m
- Structure Determination without Mass Spect47m
- Mass Spectrometry12m
- Mass Spect:Fragmentation28m
- Mass Spect:Isotopes27m
- 16. Conjugated Systems6h 13m
- Conjugation Chemistry13m
- Stability of Conjugated Intermediates4m
- Allylic Halogenation12m
- Reactions at the Allylic Position39m
- Conjugated Hydrohalogenation (1,2 vs 1,4 addition)26m
- Diels-Alder Reaction9m
- Diels-Alder Forming Bridged Products11m
- Diels-Alder Retrosynthesis8m
- Molecular Orbital Theory9m
- Drawing Atomic Orbitals6m
- Drawing Molecular Orbitals17m
- HOMO LUMO4m
- Orbital Diagram:3-atoms- Allylic Ions13m
- Orbital Diagram:4-atoms- 1,3-butadiene11m
- Orbital Diagram:5-atoms- Allylic Ions10m
- Orbital Diagram:6-atoms- 1,3,5-hexatriene13m
- Orbital Diagram:Excited States4m
- Pericyclic Reaction10m
- Thermal Cycloaddition Reactions26m
- Photochemical Cycloaddition Reactions26m
- Thermal Electrocyclic Reactions14m
- Photochemical Electrocyclic Reactions10m
- Cumulative Electrocyclic Problems25m
- Sigmatropic Rearrangement17m
- Cope Rearrangement9m
- Claisen Rearrangement15m
- 17. Ultraviolet Spectroscopy51m
- 18. Aromaticity2h 34m
- 19. Reactions of Aromatics: EAS and Beyond5h 1m
- Electrophilic Aromatic Substitution9m
- Benzene Reactions11m
- EAS:Halogenation Mechanism6m
- EAS:Nitration Mechanism9m
- EAS:Friedel-Crafts Alkylation Mechanism6m
- EAS:Friedel-Crafts Acylation Mechanism5m
- EAS:Any Carbocation Mechanism7m
- Electron Withdrawing Groups22m
- EAS:Ortho vs. Para Positions4m
- Acylation of Aniline9m
- Limitations of Friedel-Crafts Alkyation19m
- Advantages of Friedel-Crafts Acylation6m
- Blocking Groups - Sulfonic Acid12m
- EAS:Synergistic and Competitive Groups13m
- Side-Chain Halogenation6m
- Side-Chain Oxidation4m
- Reactions at Benzylic Positions31m
- Birch Reduction10m
- EAS:Sequence Groups4m
- EAS:Retrosynthesis29m
- Diazo Replacement Reactions6m
- Diazo Sequence Groups5m
- Diazo Retrosynthesis13m
- Nucleophilic Aromatic Substitution28m
- Benzyne16m
- 20. Phenols55m
- 21. Aldehydes and Ketones: Nucleophilic Addition4h 56m
- Naming Aldehydes8m
- Naming Ketones7m
- Oxidizing and Reducing Agents9m
- Oxidation of Alcohols28m
- Ozonolysis7m
- DIBAL5m
- Alkyne Hydration9m
- Nucleophilic Addition8m
- Cyanohydrin11m
- Organometallics on Ketones19m
- Overview of Nucleophilic Addition of Solvents13m
- Hydrates6m
- Hemiacetal9m
- Acetal12m
- Acetal Protecting Group16m
- Thioacetal6m
- Imine vs Enamine15m
- Addition of Amine Derivatives5m
- Wolff Kishner Reduction7m
- Baeyer-Villiger Oxidation39m
- Acid Chloride to Ketone7m
- Nitrile to Ketone9m
- Wittig Reaction18m
- Ketone and Aldehyde Synthesis Reactions14m
- 22. Carboxylic Acid Derivatives: NAS2h 51m
- Carboxylic Acid Derivatives7m
- Naming Carboxylic Acids9m
- Diacid Nomenclature6m
- Naming Esters5m
- Naming Nitriles3m
- Acid Chloride Nomenclature5m
- Naming Anhydrides7m
- Naming Amides5m
- Nucleophilic Acyl Substitution18m
- Carboxylic Acid to Acid Chloride6m
- Fischer Esterification5m
- Acid-Catalyzed Ester Hydrolysis4m
- Saponification3m
- Transesterification5m
- Lactones, Lactams and Cyclization Reactions10m
- Carboxylation5m
- Decarboxylation Mechanism14m
- Review of Nitriles46m
- 23. The Chemistry of Thioesters, Phophate Ester and Phosphate Anhydrides1h 10m
- 24. Enolate Chemistry: Reactions at the Alpha-Carbon1h 53m
- Tautomerization9m
- Tautomers of Dicarbonyl Compounds6m
- Enolate4m
- Acid-Catalyzed Alpha-Halogentation4m
- Base-Catalyzed Alpha-Halogentation3m
- Haloform Reaction8m
- Hell-Volhard-Zelinski Reaction3m
- Overview of Alpha-Alkylations and Acylations5m
- Enolate Alkylation and Acylation12m
- Enamine Alkylation and Acylation16m
- Beta-Dicarbonyl Synthesis Pathway7m
- Acetoacetic Ester Synthesis13m
- Malonic Ester Synthesis15m
- 25. Condensation Chemistry2h 9m
- 26. Amines1h 43m
- 27. Heterocycles2h 0m
- Nomenclature of Heterocycles15m
- Acid-Base Properties of Nitrogen Heterocycles10m
- Reactions of Pyrrole, Furan, and Thiophene13m
- Directing Effects in Substituted Pyrroles, Furans, and Thiophenes16m
- Addition Reactions of Furan8m
- EAS Reactions of Pyridine17m
- SNAr Reactions of Pyridine18m
- Side-Chain Reactions of Substituted Pyridines20m
- 28. Carbohydrates5h 53m
- Monosaccharide20m
- Monosaccharides - D and L Isomerism9m
- Monosaccharides - Drawing Fischer Projections18m
- Monosaccharides - Common Structures6m
- Monosaccharides - Forming Cyclic Hemiacetals12m
- Monosaccharides - Cyclization18m
- Monosaccharides - Haworth Projections13m
- Mutarotation11m
- Epimerization9m
- Monosaccharides - Aldose-Ketose Rearrangement8m
- Monosaccharides - Alkylation10m
- Monosaccharides - Acylation7m
- Glycoside6m
- Monosaccharides - N-Glycosides18m
- Monosaccharides - Reduction (Alditols)12m
- Monosaccharides - Weak Oxidation (Aldonic Acid)7m
- Reducing Sugars23m
- Monosaccharides - Strong Oxidation (Aldaric Acid)11m
- Monosaccharides - Oxidative Cleavage27m
- Monosaccharides - Osazones10m
- Monosaccharides - Kiliani-Fischer23m
- Monosaccharides - Wohl Degradation12m
- Monosaccharides - Ruff Degradation12m
- Disaccharide30m
- Polysaccharide11m
- 29. Amino Acids3h 20m
- Proteins and Amino Acids19m
- L and D Amino Acids14m
- Polar Amino Acids14m
- Amino Acid Chart18m
- Acid-Base Properties of Amino Acids33m
- Isoelectric Point14m
- Amino Acid Synthesis: HVZ Method12m
- Synthesis of Amino Acids: Acetamidomalonic Ester Synthesis16m
- Synthesis of Amino Acids: N-Phthalimidomalonic Ester Synthesis13m
- Synthesis of Amino Acids: Strecker Synthesis13m
- Reactions of Amino Acids: Esterification7m
- Reactions of Amino Acids: Acylation3m
- Reactions of Amino Acids: Hydrogenolysis6m
- Reactions of Amino Acids: Ninhydrin Test11m
- 30. Peptides and Proteins2h 42m
- Peptides12m
- Primary Protein Structure4m
- Secondary Protein Structure17m
- Tertiary Protein Structure11m
- Disulfide Bonds17m
- Quaternary Protein Structure10m
- Summary of Protein Structure7m
- Intro to Peptide Sequencing2m
- Peptide Sequencing: Partial Hydrolysis25m
- Peptide Sequencing: Partial Hydrolysis with Cyanogen Bromide7m
- Peptide Sequencing: Edman Degradation28m
- Merrifield Solid-Phase Peptide Synthesis18m
- 32. Lipids 2h 50m
- 34. Nucleic Acids1h 32m
- 35. Transition Metals5h 33m
- Electron Configuration of Elements45m
- Coordination Complexes20m
- Ligands24m
- Electron Counting10m
- The 18 and 16 Electron Rule13m
- Cross-Coupling General Reactions40m
- Heck Reaction40m
- Stille Reaction13m
- Suzuki Reaction25m
- Sonogashira Coupling Reaction17m
- Fukuyama Coupling Reaction15m
- Kumada Coupling Reaction13m
- Negishi Coupling Reaction16m
- Buchwald-Hartwig Amination Reaction19m
- Eglinton Reaction17m
- 36. Synthetic Polymers1h 49m
- Introduction to Polymers6m
- Chain-Growth Polymers10m
- Radical Polymerization15m
- Cationic Polymerization8m
- Anionic Polymerization8m
- Polymer Stereochemistry3m
- Ziegler-Natta Polymerization4m
- Copolymers6m
- Step-Growth Polymers11m
- Step-Growth Polymers: Urethane6m
- Step-Growth Polymers: Polyurethane Mechanism10m
- Step-Growth Polymers: Epoxy Resin8m
- Polymers Structure and Properties8m
Orbital Diagram:3-atoms- Allylic Ions - Online Tutor, Practice Problems & Exam Prep
 Created using AI
Created using AIIn a three-atom conjugated system, the allylic position, adjacent to a double bond, can resonate, allowing for molecular orbital explanations of reactivity. The molecular orbital theory indicates that the bonding molecular orbital (psi 1) holds the most stable electrons, while the non-bonding orbital (psi 2) has a node at the allylic position, preventing reactivity there. This illustrates why resonance structures cannot depict movement to the middle carbon, emphasizing the importance of understanding molecular orbitals in predicting chemical behavior.
Let's see what makes 3-atom conjugated systems unique from a molecular orbital perspective.
Orbital Diagram
Video transcript

Explaining Resonance through MO Theory
Video transcript
Use both resonance theory and MO theory to predict the reactive sites of the following radical. So guys, first of all, let's do the resonance one because that's the one that we should know the best. You should already know this from prior lessons. So, how does a radical resonate? Do you remember, does it resonate with 1 arrow, 2 arrows, or 3 arrows? 3. Three of the fish hook arrows. So what we would expect is that you have a resonance structure that looks like this. Fish hook, fish hook, fish hook. Does that ring a bell? So let's go ahead and draw our resonance structure and what we would expect is that now I would get a new double bond here and a new radical here. Makes sense? Cool. We would put this in brackets and then using resonance theory, which would be the reactive sites of that radical? Where would be the places that the radical could react? It would be here and here. Okay, those are the 2 positions that can react as a radical. So if we were to call this atom A, atom B, and atom C of the pi conjugated system, right, of the conjugated system, what we would say is that resonance predicts radical character at positions A and C. Right? Resonance predicts, there's an s there if you can't see it, radical character at positions A and C. Those are the positions that could react as radicals.
Now let's go ahead and actually draw out the MO diagram and see if the MO diagram also predicts it. So, how many atomic orbitals do we have? 3, right? Because we have 3 atoms that are conjugated, that means I'm going to draw 3 energy states. So this is going to be 1, 2, 3. Let me try to make it a little bit more even. There we go. Okay. So we know this is going to be Psi 1, Psi 2, Psi 3. And how many electrons in total are there? There are 3. There's 1 electron from the radical A and then there's 1 pi electron from B and there's 1 pi electron from C. So one of these is a radical because it's not bound, it's not resonating with another lone electron, so it's called a radical, but the other ones are called pi electrons because they're inside of a pi bond. Cool. Awesome. So we have those 3 single electrons. Now we have to draw our energy states. So I'm going to try to do this quickly. And some of you may already have these memorized by now because we've done this a few times, but if not that's fine, we're going to go through the rules again. So how do we predict where our lobes are, the phases? So remember the first one, let's just draw them all with the dark facing down. Remember that your next rule says that you would then draw your first one the same in every single energy state. Remember that the next rule says that you would flip the other one every time, flip flip and then we would increase the number of nodes. So we have 0 nodes on the first one and then one node on the second, meaning that I should put a node right down the middle. And then for the last one I should put a node here and a node here, which would be 2. Lastly, we have one node that needs to be deleted because I'm sorry, one orbital needs to be deleted because a node passes through it. And there you go, that's our MO diagram. Now we just have to fill it in with electrons. And what we would do is according to Aufbau principle start with and then Pauli exclusion says you can only put 2, so we're going to have to put another one up here. So what does this MO diagram tell us? What it tells us is that the radical is only going to react in 2 places and not 3. It's only going to react at orbital A and at orbital C, but it cannot react at orbital B because orbital B is a node. So, by definition, mathematically, no electrons can react at that position because it can't have any electrons. Isn't that cool? So MO theory predicts that the radical can only react at positions A and C but not B. I'm just being extra clear there because, that is a unique learning that you see once you draw the molecular orbital diagram. And really guys, when it comes down to it, Molecular Orbital Theory is the underlying theory that provides us with the knowledge of reactivity of lots of reactions. Resonance was one way to, to kind of, it's a shorthand to draw it, but really the true theory lies in the MO theory. So it's really cool that I'm getting to show this and getting to share this with you guys, and hopefully help you understand it.
Cool. So that's it for this video, let's move on to the next one.

Consider the MO's of following allyl cation. Which of the following are HOMO and LUMO?
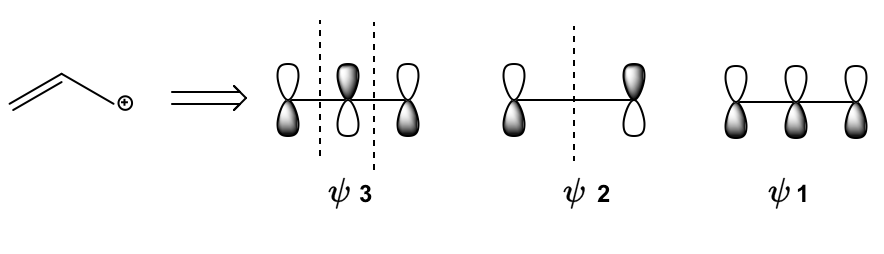
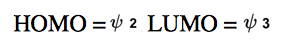




Do you want more practice?
More setsHere’s what students ask on this topic:
What is an allylic position in a three-atom conjugated system?
An allylic position in a three-atom conjugated system refers to the position adjacent to a double bond. In such systems, the allylic position is crucial because it can participate in resonance, allowing the electrons to delocalize over the system. This delocalization can be explained using molecular orbital theory, where the allylic position can resonate from one side of the double bond to the other, but not to the middle carbon due to the presence of a node in the non-bonding orbital (ψ2).
 Created using AI
Created using AIHow does molecular orbital theory explain the reactivity of allylic ions?
Molecular orbital theory explains the reactivity of allylic ions by describing the distribution of electrons in molecular orbitals. In a three-atom conjugated system, the bonding molecular orbital (ψ1) holds the most stable electrons, while the non-bonding orbital (ψ2) has a node at the middle carbon (B). This node means that electrons do not pass through this position, preventing reactivity there. As a result, the allylic ion can only react at the terminal positions (A or C), aligning with the observed resonance behavior.
 Created using AI
Created using AIWhy can't the allylic position resonate to the middle carbon in a three-atom conjugated system?
The allylic position cannot resonate to the middle carbon in a three-atom conjugated system because the non-bonding molecular orbital (ψ2) has a node at the middle carbon (B). This node indicates that no electrons are present at this position, preventing any reactivity or resonance there. Consequently, the allylic ion can only resonate between the terminal positions (A and C), but never to the middle carbon.
 Created using AI
Created using AIWhat is the significance of nodes in molecular orbitals for allylic ions?
Nodes in molecular orbitals are regions where the probability of finding an electron is zero. For allylic ions in a three-atom conjugated system, the non-bonding orbital (ψ2) has a node at the middle carbon (B). This node is significant because it prevents any reactivity at this position, as no electrons can be found there. This explains why the allylic ion can only react at the terminal positions (A or C) and not at the middle carbon, aligning with the observed resonance structures.
 Created using AI
Created using AIHow do you draw a molecular orbital diagram for a three-atom conjugated system?
To draw a molecular orbital diagram for a three-atom conjugated system, follow these steps: 1) Identify the atomic orbitals involved (A, B, and C). 2) Combine these orbitals to form molecular orbitals: bonding (ψ1), non-bonding (ψ2), and anti-bonding (ψ3). 3) Fill the molecular orbitals with electrons according to the Aufbau principle, starting with the lowest energy orbital (ψ1). 4) Note that ψ2 has a node at the middle carbon (B), preventing reactivity there. This diagram helps explain the resonance behavior and reactivity of the allylic ion.
 Created using AI
Created using AI
