Up to this point, we've been representing amino acids as these neutral structures. But it turns out that at physiological pH, meaning the pH inside your body, these amino acids don't usually look neutral. They're usually charged. So how are we going to know what types of charges to place on them? Well, that's what we're going to focus on in this video when we talk about the acid and base properties of amino acids. So at physiological pH, which is around, you know, people say 7, but it's actually like 7.4. If you're at pH 7, you're going to be in the hospital. That's not good. You need to be a little bit basic. 7.4 is the body's pH. And at that pH, amino acids exist in this state called zwitterions. You've probably heard the word zwitterion before, maybe not. But what it means is just it's a net neutral molecule with separation of charges. So let's write that in, separation of charges. So that means that, overall, the net charge is 0 but there's a positive and a negative or multiple and multiple negatives that all cancel out. Okay? So why does it exist like that? Because it's counterintuitive. Like this is the actual equilibrium, what it looks like. The amino acid starts off the way that we drew it, that we've been drawing it this whole time. It has a regular carboxylic acid. It has a regular amine. And then at physiological pH, the equilibrium shifts so that you get an anion on the O and a cation on the positive. If you think about it from a chemistry perspective, that doesn't make a whole lot of sense at first because you're always taught to think that the neutral structure is the most stable. So wouldn't it be more stable for it to stay over in that side than for it to spontaneously turn into this? Why would charges be a good thing? Why would it want to look like this? Well, the explanation goes back all the way back to our acid-base chemistry in inorganic chemistry. In a topic called determining acid-base equilibrium, we learned that the side of an equilibrium that is favored is the one with the highest pKa. We learned if you have an acid-base reaction, the side with the acid with the highest pKa is going to be the weakest acid and that's going to be the one that's the most favored. Well, that's what's happening here because it turns out that all that's happening is that this hydrogen, which I'm highlighting in red and I'm also going to highlight in yellow, is moving locations. It's getting off of the O and instead, it's joining this nitrogen as being one of the 3 Hs on that nitrogen. So it's basically moving over, okay? If you were to think of it as a mechanism, the N is actually grabbing the H and giving electrons to the O. Does that kind of make sense? Well, why would that make sense? Well because that hydrogen has to choose which acid it wants to be on. Does it want to be on a carboxylic acid, which is a pretty moderately decently strong acid? Or does it want to be on ammonium, which isn't as strong of an acid? Going back to organic chemistry, do you remember the pKa that we memorized for carboxylic acids? It was around 5, okay? And it turns out that amino acids are a little bit more acidic because they have extra groups around like nitrogen that stabilize the anion. So actually, the pKa of most carboxylic acids in amino acids is around 2. So let's write that in. The pKa is roughly 2 of that hydrogen, okay? Once that hydrogen moves over and joins the nitrogen, we learned in organic chemistry 1 that a nitrogen with a positive charge tends to have a pKa of around 10, okay? And actually, it turns out that we are very close. It's usually around 9 for amino acids. So, basically, the hydrogen has a choice. Does it stay on the acid that has a pKa of 2, or does it move to the acid with a pKa of 9? And the answer is clear from our acid-base equilibrium video, you pick the side with the highest pKa. So that's why the hydrogen doesn't want to be over here. It prefers to be on the acid with the highest pKa because that's going to be the one that's the most favored. It's the weakest acid. So that is why that's the chemistry explanation for why amino acids exist in zwitterions. Okay? So how are we going to know what types of zwitterions to draw? And it turns out that differences in the pH will actually change the charges. So how are we going to do that? We're going to find out in the next video when we talk about predominant forms.
- 1. A Review of General Chemistry5h 5m
- Summary23m
- Intro to Organic Chemistry5m
- Atomic Structure16m
- Wave Function9m
- Molecular Orbitals17m
- Sigma and Pi Bonds9m
- Octet Rule12m
- Bonding Preferences12m
- Formal Charges6m
- Skeletal Structure14m
- Lewis Structure20m
- Condensed Structural Formula15m
- Degrees of Unsaturation15m
- Constitutional Isomers14m
- Resonance Structures46m
- Hybridization23m
- Molecular Geometry16m
- Electronegativity22m
- 2. Molecular Representations1h 14m
- 3. Acids and Bases2h 46m
- 4. Alkanes and Cycloalkanes4h 19m
- IUPAC Naming29m
- Alkyl Groups13m
- Naming Cycloalkanes10m
- Naming Bicyclic Compounds10m
- Naming Alkyl Halides7m
- Naming Alkenes3m
- Naming Alcohols8m
- Naming Amines15m
- Cis vs Trans21m
- Conformational Isomers13m
- Newman Projections14m
- Drawing Newman Projections16m
- Barrier To Rotation7m
- Ring Strain8m
- Axial vs Equatorial7m
- Cis vs Trans Conformations4m
- Equatorial Preference14m
- Chair Flip9m
- Calculating Energy Difference Between Chair Conformations17m
- A-Values17m
- Decalin7m
- 5. Chirality3h 39m
- Constitutional Isomers vs. Stereoisomers9m
- Chirality12m
- Test 1:Plane of Symmetry7m
- Test 2:Stereocenter Test17m
- R and S Configuration43m
- Enantiomers vs. Diastereomers13m
- Atropisomers9m
- Meso Compound12m
- Test 3:Disubstituted Cycloalkanes13m
- What is the Relationship Between Isomers?16m
- Fischer Projection10m
- R and S of Fischer Projections7m
- Optical Activity5m
- Enantiomeric Excess20m
- Calculations with Enantiomeric Percentages11m
- Non-Carbon Chiral Centers8m
- 6. Thermodynamics and Kinetics1h 22m
- 7. Substitution Reactions1h 48m
- 8. Elimination Reactions2h 30m
- 9. Alkenes and Alkynes2h 9m
- 10. Addition Reactions3h 18m
- Addition Reaction6m
- Markovnikov5m
- Hydrohalogenation6m
- Acid-Catalyzed Hydration17m
- Oxymercuration15m
- Hydroboration26m
- Hydrogenation6m
- Halogenation6m
- Halohydrin12m
- Carbene12m
- Epoxidation8m
- Epoxide Reactions9m
- Dihydroxylation8m
- Ozonolysis7m
- Ozonolysis Full Mechanism24m
- Oxidative Cleavage3m
- Alkyne Oxidative Cleavage6m
- Alkyne Hydrohalogenation3m
- Alkyne Halogenation2m
- Alkyne Hydration6m
- Alkyne Hydroboration2m
- 11. Radical Reactions1h 58m
- 12. Alcohols, Ethers, Epoxides and Thiols2h 42m
- Alcohol Nomenclature4m
- Naming Ethers6m
- Naming Epoxides18m
- Naming Thiols11m
- Alcohol Synthesis7m
- Leaving Group Conversions - Using HX11m
- Leaving Group Conversions - SOCl2 and PBr313m
- Leaving Group Conversions - Sulfonyl Chlorides7m
- Leaving Group Conversions Summary4m
- Williamson Ether Synthesis3m
- Making Ethers - Alkoxymercuration4m
- Making Ethers - Alcohol Condensation4m
- Making Ethers - Acid-Catalyzed Alkoxylation4m
- Making Ethers - Cumulative Practice10m
- Ether Cleavage8m
- Alcohol Protecting Groups3m
- t-Butyl Ether Protecting Groups5m
- Silyl Ether Protecting Groups10m
- Sharpless Epoxidation9m
- Thiol Reactions6m
- Sulfide Oxidation4m
- 13. Alcohols and Carbonyl Compounds2h 17m
- 14. Synthetic Techniques1h 26m
- 15. Analytical Techniques:IR, NMR, Mass Spect7h 3m
- Purpose of Analytical Techniques5m
- Infrared Spectroscopy16m
- Infrared Spectroscopy Table31m
- IR Spect:Drawing Spectra40m
- IR Spect:Extra Practice26m
- NMR Spectroscopy10m
- 1H NMR:Number of Signals26m
- 1H NMR:Q-Test26m
- 1H NMR:E/Z Diastereoisomerism8m
- H NMR Table24m
- 1H NMR:Spin-Splitting (N + 1) Rule22m
- 1H NMR:Spin-Splitting Simple Tree Diagrams11m
- 1H NMR:Spin-Splitting Complex Tree Diagrams12m
- 1H NMR:Spin-Splitting Patterns8m
- NMR Integration18m
- NMR Practice14m
- Carbon NMR4m
- Structure Determination without Mass Spect47m
- Mass Spectrometry12m
- Mass Spect:Fragmentation28m
- Mass Spect:Isotopes27m
- 16. Conjugated Systems6h 13m
- Conjugation Chemistry13m
- Stability of Conjugated Intermediates4m
- Allylic Halogenation12m
- Reactions at the Allylic Position39m
- Conjugated Hydrohalogenation (1,2 vs 1,4 addition)26m
- Diels-Alder Reaction9m
- Diels-Alder Forming Bridged Products11m
- Diels-Alder Retrosynthesis8m
- Molecular Orbital Theory9m
- Drawing Atomic Orbitals6m
- Drawing Molecular Orbitals17m
- HOMO LUMO4m
- Orbital Diagram:3-atoms- Allylic Ions13m
- Orbital Diagram:4-atoms- 1,3-butadiene11m
- Orbital Diagram:5-atoms- Allylic Ions10m
- Orbital Diagram:6-atoms- 1,3,5-hexatriene13m
- Orbital Diagram:Excited States4m
- Pericyclic Reaction10m
- Thermal Cycloaddition Reactions26m
- Photochemical Cycloaddition Reactions26m
- Thermal Electrocyclic Reactions14m
- Photochemical Electrocyclic Reactions10m
- Cumulative Electrocyclic Problems25m
- Sigmatropic Rearrangement17m
- Cope Rearrangement9m
- Claisen Rearrangement15m
- 17. Ultraviolet Spectroscopy51m
- 18. Aromaticity2h 34m
- 19. Reactions of Aromatics: EAS and Beyond5h 1m
- Electrophilic Aromatic Substitution9m
- Benzene Reactions11m
- EAS:Halogenation Mechanism6m
- EAS:Nitration Mechanism9m
- EAS:Friedel-Crafts Alkylation Mechanism6m
- EAS:Friedel-Crafts Acylation Mechanism5m
- EAS:Any Carbocation Mechanism7m
- Electron Withdrawing Groups22m
- EAS:Ortho vs. Para Positions4m
- Acylation of Aniline9m
- Limitations of Friedel-Crafts Alkyation19m
- Advantages of Friedel-Crafts Acylation6m
- Blocking Groups - Sulfonic Acid12m
- EAS:Synergistic and Competitive Groups13m
- Side-Chain Halogenation6m
- Side-Chain Oxidation4m
- Reactions at Benzylic Positions31m
- Birch Reduction10m
- EAS:Sequence Groups4m
- EAS:Retrosynthesis29m
- Diazo Replacement Reactions6m
- Diazo Sequence Groups5m
- Diazo Retrosynthesis13m
- Nucleophilic Aromatic Substitution28m
- Benzyne16m
- 20. Phenols55m
- 21. Aldehydes and Ketones: Nucleophilic Addition4h 56m
- Naming Aldehydes8m
- Naming Ketones7m
- Oxidizing and Reducing Agents9m
- Oxidation of Alcohols28m
- Ozonolysis7m
- DIBAL5m
- Alkyne Hydration9m
- Nucleophilic Addition8m
- Cyanohydrin11m
- Organometallics on Ketones19m
- Overview of Nucleophilic Addition of Solvents13m
- Hydrates6m
- Hemiacetal9m
- Acetal12m
- Acetal Protecting Group16m
- Thioacetal6m
- Imine vs Enamine15m
- Addition of Amine Derivatives5m
- Wolff Kishner Reduction7m
- Baeyer-Villiger Oxidation39m
- Acid Chloride to Ketone7m
- Nitrile to Ketone9m
- Wittig Reaction18m
- Ketone and Aldehyde Synthesis Reactions14m
- 22. Carboxylic Acid Derivatives: NAS2h 51m
- Carboxylic Acid Derivatives7m
- Naming Carboxylic Acids9m
- Diacid Nomenclature6m
- Naming Esters5m
- Naming Nitriles3m
- Acid Chloride Nomenclature5m
- Naming Anhydrides7m
- Naming Amides5m
- Nucleophilic Acyl Substitution18m
- Carboxylic Acid to Acid Chloride6m
- Fischer Esterification5m
- Acid-Catalyzed Ester Hydrolysis4m
- Saponification3m
- Transesterification5m
- Lactones, Lactams and Cyclization Reactions10m
- Carboxylation5m
- Decarboxylation Mechanism14m
- Review of Nitriles46m
- 23. The Chemistry of Thioesters, Phophate Ester and Phosphate Anhydrides1h 10m
- 24. Enolate Chemistry: Reactions at the Alpha-Carbon1h 53m
- Tautomerization9m
- Tautomers of Dicarbonyl Compounds6m
- Enolate4m
- Acid-Catalyzed Alpha-Halogentation4m
- Base-Catalyzed Alpha-Halogentation3m
- Haloform Reaction8m
- Hell-Volhard-Zelinski Reaction3m
- Overview of Alpha-Alkylations and Acylations5m
- Enolate Alkylation and Acylation12m
- Enamine Alkylation and Acylation16m
- Beta-Dicarbonyl Synthesis Pathway7m
- Acetoacetic Ester Synthesis13m
- Malonic Ester Synthesis15m
- 25. Condensation Chemistry2h 9m
- 26. Amines1h 43m
- 27. Heterocycles2h 0m
- Nomenclature of Heterocycles15m
- Acid-Base Properties of Nitrogen Heterocycles10m
- Reactions of Pyrrole, Furan, and Thiophene13m
- Directing Effects in Substituted Pyrroles, Furans, and Thiophenes16m
- Addition Reactions of Furan8m
- EAS Reactions of Pyridine17m
- SNAr Reactions of Pyridine18m
- Side-Chain Reactions of Substituted Pyridines20m
- 28. Carbohydrates5h 53m
- Monosaccharide20m
- Monosaccharides - D and L Isomerism9m
- Monosaccharides - Drawing Fischer Projections18m
- Monosaccharides - Common Structures6m
- Monosaccharides - Forming Cyclic Hemiacetals12m
- Monosaccharides - Cyclization18m
- Monosaccharides - Haworth Projections13m
- Mutarotation11m
- Epimerization9m
- Monosaccharides - Aldose-Ketose Rearrangement8m
- Monosaccharides - Alkylation10m
- Monosaccharides - Acylation7m
- Glycoside6m
- Monosaccharides - N-Glycosides18m
- Monosaccharides - Reduction (Alditols)12m
- Monosaccharides - Weak Oxidation (Aldonic Acid)7m
- Reducing Sugars23m
- Monosaccharides - Strong Oxidation (Aldaric Acid)11m
- Monosaccharides - Oxidative Cleavage27m
- Monosaccharides - Osazones10m
- Monosaccharides - Kiliani-Fischer23m
- Monosaccharides - Wohl Degradation12m
- Monosaccharides - Ruff Degradation12m
- Disaccharide30m
- Polysaccharide11m
- 29. Amino Acids3h 20m
- Proteins and Amino Acids19m
- L and D Amino Acids14m
- Polar Amino Acids14m
- Amino Acid Chart18m
- Acid-Base Properties of Amino Acids33m
- Isoelectric Point14m
- Amino Acid Synthesis: HVZ Method12m
- Synthesis of Amino Acids: Acetamidomalonic Ester Synthesis16m
- Synthesis of Amino Acids: N-Phthalimidomalonic Ester Synthesis13m
- Synthesis of Amino Acids: Strecker Synthesis13m
- Reactions of Amino Acids: Esterification7m
- Reactions of Amino Acids: Acylation3m
- Reactions of Amino Acids: Hydrogenolysis6m
- Reactions of Amino Acids: Ninhydrin Test11m
- 30. Peptides and Proteins2h 42m
- Peptides12m
- Primary Protein Structure4m
- Secondary Protein Structure17m
- Tertiary Protein Structure11m
- Disulfide Bonds17m
- Quaternary Protein Structure10m
- Summary of Protein Structure7m
- Intro to Peptide Sequencing2m
- Peptide Sequencing: Partial Hydrolysis25m
- Peptide Sequencing: Partial Hydrolysis with Cyanogen Bromide7m
- Peptide Sequencing: Edman Degradation28m
- Merrifield Solid-Phase Peptide Synthesis18m
- 31. Catalysis in Organic Reactions1h 30m
- 32. Lipids 2h 50m
- 34. Nucleic Acids1h 32m
- 35. Transition Metals5h 33m
- Electron Configuration of Elements45m
- Coordination Complexes20m
- Ligands24m
- Electron Counting10m
- The 18 and 16 Electron Rule13m
- Cross-Coupling General Reactions40m
- Heck Reaction40m
- Stille Reaction13m
- Suzuki Reaction25m
- Sonogashira Coupling Reaction17m
- Fukuyama Coupling Reaction15m
- Kumada Coupling Reaction13m
- Negishi Coupling Reaction16m
- Buchwald-Hartwig Amination Reaction19m
- Eglinton Reaction17m
- 36. Synthetic Polymers1h 49m
- Introduction to Polymers6m
- Chain-Growth Polymers10m
- Radical Polymerization15m
- Cationic Polymerization8m
- Anionic Polymerization8m
- Polymer Stereochemistry3m
- Ziegler-Natta Polymerization4m
- Copolymers6m
- Step-Growth Polymers11m
- Step-Growth Polymers: Urethane6m
- Step-Growth Polymers: Polyurethane Mechanism10m
- Step-Growth Polymers: Epoxy Resin8m
- Polymers Structure and Properties8m
Acid-Base Properties of Amino Acids: Study with Video Lessons, Practice Problems & Examples
 Created using AI
Created using AIAmino acids exist as zwitterions at physiological pH (around 7.4), featuring both positive and negative charges that balance to a net charge of zero. Their amphoteric nature allows them to act as acids or bases depending on the pH, influenced by their pKa values. For instance, at pH levels below the pKa of the carboxylic acid (approximately 2), amino acids become protonated, while at pH levels above the pKa of the amine (around 9), they lose protons. Additionally, seven amino acids possess ionizable side chains, requiring consideration of their pKa values for accurate charge determination.
Amino acids aren't always represented as neutral structures. It's time to represent them appropriately.
At physiological pH (7.4), amino acids exist as zwitterions. Let's take a deeper look.
Why Amino Acids Exist as Zwitterions
Video transcript

Determining Predominant Forms
Video transcript
Like water, amino acids are amphoteric, and "amphoteric" is just a fancy word for a molecule that can react either as an acid or as a base depending on the situation. And the reason I mentioned water is because water is the most famous example of an amphoteric molecule. Remember that water could either accept a proton or it could donate a proton based on its surroundings, based on the pKa and the pH of the solution. Well, the same thing is true of amino acids because amino acids have an acidic carboxylic acid group and a basic amine group. So, we're going to need to figure out when it's reacting as an acid and when it's reacting as a base. And for that, we're going to be using exact pKa values. We use exact pKa values given to us on a table to figure out at a certain pH, does that amino acid exist in a charged state or is it in a neutral state, positive or negative. Now, I've gone ahead and provided all these pKas for you in your amino acid breakdown sheet. We're not going to look at it right now, but I just want to remind you, remember all those numbers you saw next to the amino acids? This is what we're using them for. We're going to use them to determine the charges of the amino acids at different pHs, okay?
Before we look at an example, I want to remind you about the theory behind this. Remember that in general chemistry, there was an equation that we learned called the Henderson-Hasselbalch equation. Now, we're not going to have to use that equation in this lesson, but there is just this idea from it that you should be aware of, which is that when the pH of a solution is exactly equal to the pKa of an acid, then exactly half of that functional group is going to be ionized, okay? So, what that means is that if your pH and your pKa are exactly equal to the same amount, that means that that group, that functional group will exist as a 50% neutral molecule and as a 50% charged molecule, maybe positive, maybe negative. It depends on the situation. That's neutral and charged. I'm sorry, it's a little bit off the screen. So what that means is that when they're at equilibrium like that, the neutral and charged forms are even with each other. But once you make the pH a little bit lower than the pKa, what's going to happen? Well, now, you're going to get a more protonated form that predominates because now there's going to be more acid around, so you're going to get more charges, more positives. And if the pH is higher than the pKa, then it's going to be more basic in the solution, so the deprotonated form is going to predominate, okay?
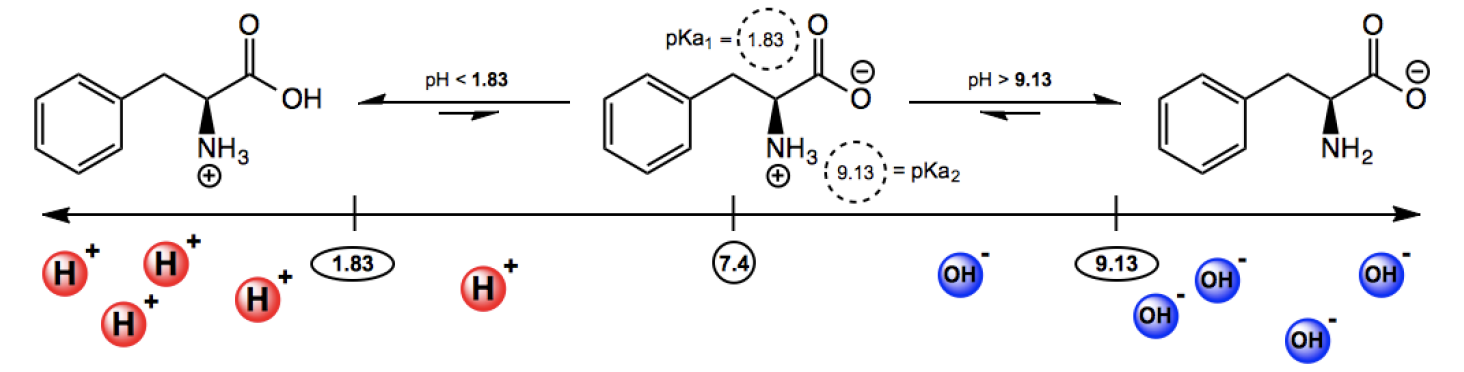
So I want to show you guys an example of what this could look like because it turns out that if you look at the pKa table for phenylalanine, the pKa1, or the pKa of the carboxylic acid group, it's not actually 2; it's close to 2, it's 1.83. You actually need to look that up, okay? You can't just say 2. The exact pKa2, or the pKa of the nitrogen, is not 9; it's 9.13, very similar but just a little bit higher. So, what that means is that at physiological pH of 7.4, like we already discussed, it's going to exist as a zwitterion. It's going to exist where the oxygen gives up its protons to the nitrogen so that the oxygen is negative, and so that the nitrogen is positive. And actually, this is going to be true for any pH all the way from 1.83 all the way to 9.13. So basically, for like 8 numbers of pH, it's going to look like this because in all these situations, the zwitterionic form is the most stable because in all these situations, the pH hasn't become a greater number than the pKa or a lesser number than the pKa.
But what starts to happen once you reduce the pH of the solution below 1.83? Well, when you think of it conceptually, if the pH is 1.83, is that acidic or basic? Super acidic, right? So, it means there's protons everywhere. So this negative charge has been holding on. It's been negative for a while. But what happens if I start slamming it with protons? Like I keep making it more and more acidic, right? What if I drop it to 1.83? What if I drop it to 1.5? What if I drop it to 1? What if I drop it to 0.5? You get what I'm saying? You keep throwing protons at that thing; eventually, it's going to get protonated. Eventually, at some point, if I add enough protons to it, I'm going to get a proton there. That's what 1.83 is. Any pH below 1.83, you add the proton. So that means that at pH 1.83 or below, the pH is less than the pKa1. pKa1 is 1.83, and that means that the protonated form predominates.
Now, the same thing with the nitrogen, so what happens if I start raising the pH of the solution all the way to like 10, all the way to 11, right? So this thing is getting super basic. There's hydroxide molecules everywhere. And at some point, one of these H? So, how do I know when the hydroxide is going to take away that H? You look at the pKa2 value. The pKa2 value is 9.13. So that means that any pH that is higher than 9.13, there's going to be so much base around that that nitrogen won't be able to hold on to that proton and eventually, it's going to lose that proton. Now, it doesn't become negatively charged, that's a different idea. But now, it used to be positive and now it goes into the neutral. And the deprotonated form, in this sense, doesn't mean that it always has a negative. And protonated doesn't always mean that it has a positive because actually, my protonated version didn't have a positive either. It just depends on the molecule you're starting with. But deprotonated means that I lost a proton, and protonated meant that I gained a proton. Does that make sense? Cool. So this is the way that we're going to be thinking about predominant forms. We're going to have to compare pKas using the sheet and then we're going to use the simple formula to figure out what form it should be in. So that's it for this video. Let's move on to the next one.
The 7 Ionizable Amino Acids
Video transcript
So you think you're getting the hang of this. This is kind of making sense. And now I'm going to throw a wrench into the whole thing. Because it turns out that there's an extra complication which is that 7 of the 20 amino acids that you learned actually also have ionizable side chains. What that means is that when you are planning out the zwitterionic form, if it has positives, negatives, etcetera, you need to also look at the side chain and you have to figure out if the side chain is going to be protonated or deprotonated, etcetera. On top of that, some professors, some universities make you memorize these 7 pKa's. So I'm going to give them to you and I don't really have the best memory tools for them. It's going to be up to you to figure out if you need to memorize these or if they're going to be given to you. Most of the time, 90% of the time, they're given to you just like the others, so you don't have to worry. But if you happen to have a professor that wants you to memorize them, then you're going to need to figure out a way to memorize them. Okay? So let's go ahead and move to the next page. I'm going to move to basically back to the amino acid breakdown page. Okay? So this is back to the amino acid breakdown and now we're going to be filling in these boxes over here and the 7 ionizable pKa's or side chains are the ones that have white boxes. So we're going to be filling in numbers here. Okay? Cool.
Another disclaimer. It turns out that not only does not everyone have to memorize them, some people do, some people don't, but there are also differing values depending on your professor, depending on what website you look at. Some will say 8.3 and some will say 8.4. It's like really stupid. So I personally don't think it's worth memorizing unless your professor makes you because no one really fully agrees. I'm going to go ahead and give you the values that I found were the most popular ones. But if your professor or textbook requires you to memorize a slightly different one, use that one. Okay? So cysteine, 8.18. I'm not going to give you ways to memorize this. I'm just going to write them down and you figure that out if you need to. Tyrosine, 10.07. Aspartic acid, 3.65. Glutamic acid, 4.25. By the way, that kind of makes sense. Right? That aspartic would be more acidic than glutamic because it's closer to the nitrogen, so it's going to have a stronger inductive effect and this one's further away. Kinda makes sense. Lysine, 10.53. Histidine is exactly 6. That one seems to be pretty universally agreed on. It's almost always 6. And then arginine, 12.48. Okay? So just I'm not going to help you memorize these. You probably don't even need to, but just have them written down so we can use them in the following problems. We're going to referring to this sheet a lot because we're going to need to get all our pKa values from here. Okay? Now, I happen to know that the next example is with Lysine. So let's get these Let's write down these numbers now so that we can use them on the next page. And I'm actually going to write them down myself so that I can use them on the next page. So lysine, the carboxylic acid is 2.18. The amine is 8.95. As you can see, again, they're close to 2.9, but they're a little bit off. And then the lysine side chain is 10.53. So now let's go over to the example, figure out what the predominant form of lysine looks like and what's the net charge at a pH of 8.5. Cool? So actually, let's just do this together. Okay? Actually, okay. No. I'll let you guys try it first, and then in the next video, I will solve it for you.
As mentioned, 7 of the 20 amino acids have ionizable sidechains. The 7 are:Cysteine (C), Tyrosine (Y), Aspartic Acid (D), Glutamic Acid (E), Lysine (K), Histadine (H), and Arginine (R).
Predicting Predominant Form of Lysine
Video transcript
So let's write in our pKa's. This one, pKa 1, was 2.18. This one was 8.95. And then finally, this one was 10.53. Also, notice that I didn't include any hydrogens because the hydrogens are going to depend on the charge. So let's just go pKa by pKa, okay? So the first one. So remember, the pH is 8.5. So for pKa 1, is pH less than or greater than 2.18? It's greater than, right? So pH is greater than pKa 1. Does that make sense? And I'm just gonna write in the numbers just so you guys can see. pH is 8.5. pKa 1 is 2.18. Which one is bigger? pH, right? So what does that mean? Well, when the pH is greater, that means deprotonated. Right? We have to look at our rules. Deprotonated predominates, right, because it's basic. So I'm gonna put here deprotonated. And for this, deprotonated means it needs a negative charge. Cool. So we figured out the first charge already. Awesome. So let's look at the next one. So is the pH greater than or less than pKa 2? It's actually less than, right? Because pKa 2 is 8.95. So in this case, pH is less than pKa 2. And once again, just to make it really clear, what I'm saying is that 8.5 is a lower number than 8.95, correct? So since it's lower, that means that it's more acidic. That means protonated predominates. Let me avoid writing over the lysine. Okay? So that means that the protonated form of this amine is NH3+. Right? So actually, we've got our zwitterion. At this pH of 8.5, we've got our zwitterion. By the way, I could have already known that because 8.5 is in between the two pKa's. Remember we said any pH that's between the two pKa's will form the zwitterion. So 8.5 makes sense that it does. Then finally, 10.53. So pH is less than pKa 3. And just to be super clear, that means that 8.5 is less than the number 10.53, right? So since it's less, that means it's more acidic which means that the protonated form should predominate. And the protonated form of the amine is NH3. So that means that this should also be NH3+. Cool. So now we figured out the predominant form at this specific pH. What is the net charge? Well, the net charge is just summing all the charges. So I have two positives and one negative. So that means that the net charge is equal to one positive. Okay? Or you could just put a bracket around and just say that it's positive. Okay? Cool. So now you guys get the fact that predominant forms aren't just with the backbone. You also may have to include the side chain and you may need to memorize your pKa's for the seven ionizable amino acids if your professor wants you to. But for right now, we'll just be looking at the reference sheet every time. Okay. So let's go ahead and move on to the next video.
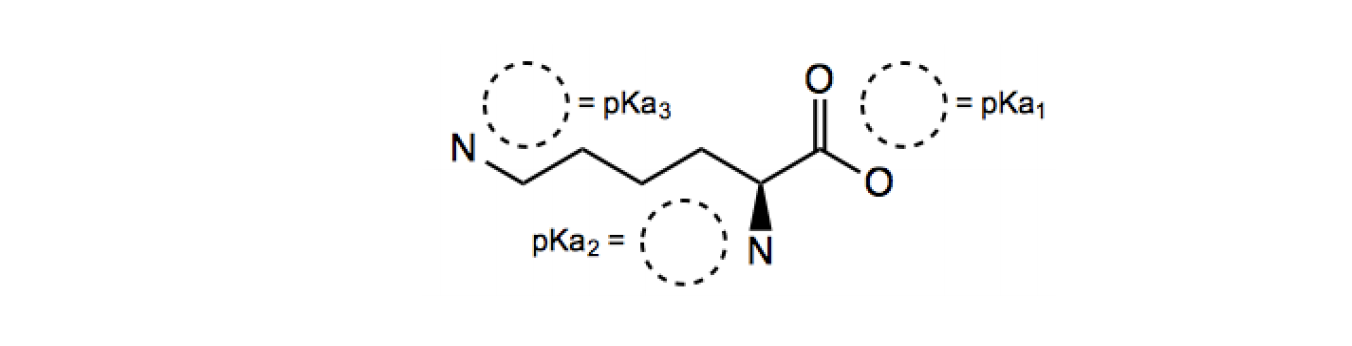
Predict the predominant form and net charge of tyrosine (Y) at pH 10. What is the net charge?
Problem Transcript
Determine the net charge of the dipeptide R-C at pH 4.3. (Hint:Peptide bonds do not count)
Problem Transcript
Do you want more practice?
More setsHere’s what students ask on this topic:
What are zwitterions and how do amino acids exist as zwitterions at physiological pH?
Zwitterions are molecules that have both positive and negative charges but are overall electrically neutral. At physiological pH (around 7.4), amino acids exist as zwitterions. This occurs because the carboxyl group (–COOH) loses a proton (H+), becoming negatively charged (–COO−), while the amino group (–NH2) gains a proton, becoming positively charged (–NH3+). The result is a molecule with separated charges that balance out to a net charge of zero. This form is more stable at physiological pH due to the relative pKa values of the carboxyl and amino groups.
 Created using AI
Created using AIHow do the pKa values of amino acids influence their charge at different pH levels?
The pKa values of amino acids determine the pH at which their functional groups (carboxyl and amino groups) gain or lose protons. When the pH is below the pKa of the carboxyl group (around 2), the carboxyl group remains protonated (–COOH). When the pH is above the pKa of the amino group (around 9), the amino group loses a proton (–NH2). At intermediate pH levels, such as physiological pH (7.4), the carboxyl group is deprotonated (–COO−) and the amino group is protonated (–NH3+), resulting in a zwitterion.
 Created using AI
Created using AIWhat is the Henderson-Hasselbalch equation and how is it used to determine the charge of amino acids?
The Henderson-Hasselbalch equation is used to relate the pH of a solution to the pKa of an acid and the ratio of the concentrations of its protonated and deprotonated forms. The equation is:
In the context of amino acids, it helps determine whether the amino acid will be in a protonated or deprotonated state at a given pH. When pH = pKa, the concentrations of the protonated and deprotonated forms are equal.
 Created using AI
Created using AIWhich amino acids have ionizable side chains and what are their pKa values?
Seven amino acids have ionizable side chains: cysteine (pKa ≈ 8.18), tyrosine (pKa ≈ 10.07), aspartic acid (pKa ≈ 3.65), glutamic acid (pKa ≈ 4.25), lysine (pKa ≈ 10.53), histidine (pKa ≈ 6.00), and arginine (pKa ≈ 12.48). These side chains can gain or lose protons depending on the pH, affecting the overall charge of the amino acid. The exact pKa values can vary slightly depending on the source, so it is important to refer to the specific values provided by your instructor or textbook.
 Created using AI
Created using AIHow does the amphoteric nature of amino acids affect their behavior in different pH environments?
Amino acids are amphoteric, meaning they can act as both acids and bases. This dual behavior is due to the presence of both an acidic carboxyl group (–COOH) and a basic amino group (–NH2). In acidic environments (low pH), amino acids tend to gain protons, becoming positively charged. In basic environments (high pH), they lose protons, becoming negatively charged. At physiological pH (around 7.4), they exist as zwitterions, with both positive and negative charges that balance out to a net charge of zero.
 Created using AI
Created using AIYour Organic Chemistry tutors
- Glycine has pKa values of 2.34 and 9.60. At what pH does glycine exist in the forms shown? a.
- Alanine has pKa values of 2.34 and 9.69. Therefore, alanine exists predominately as a zwitterion in an aqueous...
- Identify the location and type of charge on the hexapeptide Lys-Ser-Asp-Cys-His-Tyr at each of the following p...
- Identify the location and type of charge on the hexapeptide Lys-Ser-Asp-Cys-His-Tyr at each of the following p...
- Draw the form of aspartate that predominates at the following pH values: d. pH=11.0
- Draw the form of aspartate that predominates at the following pH values: c. pH=6.0
- Draw the predominant form for glutamate in a solution with the following pH: b. 3
- Draw the predominant form for glutamate in a solution with the following pH: a. 0
- c. Which amino acid has the greatest amount of negative charge at pH = 6.20?
- Draw the pH–activity profile for an enzyme that has one catalytic group at the active site: a. the catalytic ...
- Draw the pH–activity profile for an enzyme that has one catalytic group at the active site: b. the catalytic ...
- The pH–activity profile for glucose-6-phosphate isomerase indicates the participation of a group with a pKa = ...
- Draw the structure of the predominant form of (b) proline at pH 2.
- Draw the structure of the predominant form of (e) a mixture of alanine, lysine, and aspartic acid at (i) pH 6;
- Draw the structure of the predominant form of (a) isoleucine at pH 11.
- Draw the structure of the predominant form of(d) glutamic acid at pH 7.
- Draw the structure of the predominant form of(e) a mixture of alanine, lysine, and aspartic acid at (iii) pH 2...
- Although tryptophan contains a heterocyclic amine, it is considered a neutral amino acid. Explain why the indo...
- Aspartame (Nutrasweet®) is a remarkably sweet-tasting dipeptide ester. Complete hydrolysis of aspartame gives...
- (••) Draw the following amino acids in all possible protonation states as you move from high pH to low pH.b. H...
- Draw the electrophoretic separation of Ala, Lys, and Asp at pH 9.7.

