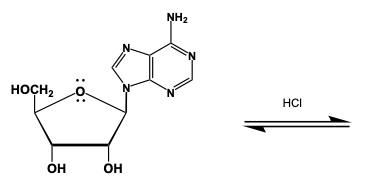
Hey, everyone. So in this video, let's talk about the hydrolysis of nucleosides. Now we're going to say that nucleosides are not easily hydrolyzed. However, given a strong enough acid and time, it's possible. Now, when we do the hydrolysis of a nucleoside, we produce as a result of this our sugar, our free sugar, and a base, our nitrogenous base. Now, here when we're looking at the acid-catalyzed hydrolysis mechanism, it can be broken down into 4 steps. Step 1 deals with a proton transfer. Step 2, we have the loss of our leaving group. Step 3, we have our nucleophilic attack. And then finally, step 4, we have yet another proton transfer. So, just remember these individual steps help to make the overall mechanism for the acidic hydrolysis of our nucleoside. And now that we've talked about it, click on the next video and let's take a look at an example question.
- 1. A Review of General Chemistry5h 5m
- Summary23m
- Intro to Organic Chemistry5m
- Atomic Structure16m
- Wave Function9m
- Molecular Orbitals17m
- Sigma and Pi Bonds9m
- Octet Rule12m
- Bonding Preferences12m
- Formal Charges6m
- Skeletal Structure14m
- Lewis Structure20m
- Condensed Structural Formula15m
- Degrees of Unsaturation15m
- Constitutional Isomers14m
- Resonance Structures46m
- Hybridization23m
- Molecular Geometry16m
- Electronegativity22m
- 2. Molecular Representations1h 14m
- 3. Acids and Bases2h 46m
- 4. Alkanes and Cycloalkanes4h 19m
- IUPAC Naming29m
- Alkyl Groups13m
- Naming Cycloalkanes10m
- Naming Bicyclic Compounds10m
- Naming Alkyl Halides7m
- Naming Alkenes3m
- Naming Alcohols8m
- Naming Amines15m
- Cis vs Trans21m
- Conformational Isomers13m
- Newman Projections14m
- Drawing Newman Projections16m
- Barrier To Rotation7m
- Ring Strain8m
- Axial vs Equatorial7m
- Cis vs Trans Conformations4m
- Equatorial Preference14m
- Chair Flip9m
- Calculating Energy Difference Between Chair Conformations17m
- A-Values17m
- Decalin7m
- 5. Chirality3h 39m
- Constitutional Isomers vs. Stereoisomers9m
- Chirality12m
- Test 1:Plane of Symmetry7m
- Test 2:Stereocenter Test17m
- R and S Configuration43m
- Enantiomers vs. Diastereomers13m
- Atropisomers9m
- Meso Compound12m
- Test 3:Disubstituted Cycloalkanes13m
- What is the Relationship Between Isomers?16m
- Fischer Projection10m
- R and S of Fischer Projections7m
- Optical Activity5m
- Enantiomeric Excess20m
- Calculations with Enantiomeric Percentages11m
- Non-Carbon Chiral Centers8m
- 6. Thermodynamics and Kinetics1h 22m
- 7. Substitution Reactions1h 48m
- 8. Elimination Reactions2h 30m
- 9. Alkenes and Alkynes2h 9m
- 10. Addition Reactions3h 18m
- Addition Reaction6m
- Markovnikov5m
- Hydrohalogenation6m
- Acid-Catalyzed Hydration17m
- Oxymercuration15m
- Hydroboration26m
- Hydrogenation6m
- Halogenation6m
- Halohydrin12m
- Carbene12m
- Epoxidation8m
- Epoxide Reactions9m
- Dihydroxylation8m
- Ozonolysis7m
- Ozonolysis Full Mechanism24m
- Oxidative Cleavage3m
- Alkyne Oxidative Cleavage6m
- Alkyne Hydrohalogenation3m
- Alkyne Halogenation2m
- Alkyne Hydration6m
- Alkyne Hydroboration2m
- 11. Radical Reactions1h 58m
- 12. Alcohols, Ethers, Epoxides and Thiols2h 42m
- Alcohol Nomenclature4m
- Naming Ethers6m
- Naming Epoxides18m
- Naming Thiols11m
- Alcohol Synthesis7m
- Leaving Group Conversions - Using HX11m
- Leaving Group Conversions - SOCl2 and PBr313m
- Leaving Group Conversions - Sulfonyl Chlorides7m
- Leaving Group Conversions Summary4m
- Williamson Ether Synthesis3m
- Making Ethers - Alkoxymercuration4m
- Making Ethers - Alcohol Condensation4m
- Making Ethers - Acid-Catalyzed Alkoxylation4m
- Making Ethers - Cumulative Practice10m
- Ether Cleavage8m
- Alcohol Protecting Groups3m
- t-Butyl Ether Protecting Groups5m
- Silyl Ether Protecting Groups10m
- Sharpless Epoxidation9m
- Thiol Reactions6m
- Sulfide Oxidation4m
- 13. Alcohols and Carbonyl Compounds2h 17m
- 14. Synthetic Techniques1h 26m
- 15. Analytical Techniques:IR, NMR, Mass Spect7h 3m
- Purpose of Analytical Techniques5m
- Infrared Spectroscopy16m
- Infrared Spectroscopy Table31m
- IR Spect:Drawing Spectra40m
- IR Spect:Extra Practice26m
- NMR Spectroscopy10m
- 1H NMR:Number of Signals26m
- 1H NMR:Q-Test26m
- 1H NMR:E/Z Diastereoisomerism8m
- H NMR Table24m
- 1H NMR:Spin-Splitting (N + 1) Rule22m
- 1H NMR:Spin-Splitting Simple Tree Diagrams11m
- 1H NMR:Spin-Splitting Complex Tree Diagrams12m
- 1H NMR:Spin-Splitting Patterns8m
- NMR Integration18m
- NMR Practice14m
- Carbon NMR4m
- Structure Determination without Mass Spect47m
- Mass Spectrometry12m
- Mass Spect:Fragmentation28m
- Mass Spect:Isotopes27m
- 16. Conjugated Systems6h 13m
- Conjugation Chemistry13m
- Stability of Conjugated Intermediates4m
- Allylic Halogenation12m
- Reactions at the Allylic Position39m
- Conjugated Hydrohalogenation (1,2 vs 1,4 addition)26m
- Diels-Alder Reaction9m
- Diels-Alder Forming Bridged Products11m
- Diels-Alder Retrosynthesis8m
- Molecular Orbital Theory9m
- Drawing Atomic Orbitals6m
- Drawing Molecular Orbitals17m
- HOMO LUMO4m
- Orbital Diagram:3-atoms- Allylic Ions13m
- Orbital Diagram:4-atoms- 1,3-butadiene11m
- Orbital Diagram:5-atoms- Allylic Ions10m
- Orbital Diagram:6-atoms- 1,3,5-hexatriene13m
- Orbital Diagram:Excited States4m
- Pericyclic Reaction10m
- Thermal Cycloaddition Reactions26m
- Photochemical Cycloaddition Reactions26m
- Thermal Electrocyclic Reactions14m
- Photochemical Electrocyclic Reactions10m
- Cumulative Electrocyclic Problems25m
- Sigmatropic Rearrangement17m
- Cope Rearrangement9m
- Claisen Rearrangement15m
- 17. Ultraviolet Spectroscopy51m
- 18. Aromaticity2h 34m
- 19. Reactions of Aromatics: EAS and Beyond5h 1m
- Electrophilic Aromatic Substitution9m
- Benzene Reactions11m
- EAS:Halogenation Mechanism6m
- EAS:Nitration Mechanism9m
- EAS:Friedel-Crafts Alkylation Mechanism6m
- EAS:Friedel-Crafts Acylation Mechanism5m
- EAS:Any Carbocation Mechanism7m
- Electron Withdrawing Groups22m
- EAS:Ortho vs. Para Positions4m
- Acylation of Aniline9m
- Limitations of Friedel-Crafts Alkyation19m
- Advantages of Friedel-Crafts Acylation6m
- Blocking Groups - Sulfonic Acid12m
- EAS:Synergistic and Competitive Groups13m
- Side-Chain Halogenation6m
- Side-Chain Oxidation4m
- Reactions at Benzylic Positions31m
- Birch Reduction10m
- EAS:Sequence Groups4m
- EAS:Retrosynthesis29m
- Diazo Replacement Reactions6m
- Diazo Sequence Groups5m
- Diazo Retrosynthesis13m
- Nucleophilic Aromatic Substitution28m
- Benzyne16m
- 20. Phenols55m
- 21. Aldehydes and Ketones: Nucleophilic Addition4h 56m
- Naming Aldehydes8m
- Naming Ketones7m
- Oxidizing and Reducing Agents9m
- Oxidation of Alcohols28m
- Ozonolysis7m
- DIBAL5m
- Alkyne Hydration9m
- Nucleophilic Addition8m
- Cyanohydrin11m
- Organometallics on Ketones19m
- Overview of Nucleophilic Addition of Solvents13m
- Hydrates6m
- Hemiacetal9m
- Acetal12m
- Acetal Protecting Group16m
- Thioacetal6m
- Imine vs Enamine15m
- Addition of Amine Derivatives5m
- Wolff Kishner Reduction7m
- Baeyer-Villiger Oxidation39m
- Acid Chloride to Ketone7m
- Nitrile to Ketone9m
- Wittig Reaction18m
- Ketone and Aldehyde Synthesis Reactions14m
- 22. Carboxylic Acid Derivatives: NAS2h 51m
- Carboxylic Acid Derivatives7m
- Naming Carboxylic Acids9m
- Diacid Nomenclature6m
- Naming Esters5m
- Naming Nitriles3m
- Acid Chloride Nomenclature5m
- Naming Anhydrides7m
- Naming Amides5m
- Nucleophilic Acyl Substitution18m
- Carboxylic Acid to Acid Chloride6m
- Fischer Esterification5m
- Acid-Catalyzed Ester Hydrolysis4m
- Saponification3m
- Transesterification5m
- Lactones, Lactams and Cyclization Reactions10m
- Carboxylation5m
- Decarboxylation Mechanism14m
- Review of Nitriles46m
- 23. The Chemistry of Thioesters, Phophate Ester and Phosphate Anhydrides1h 10m
- 24. Enolate Chemistry: Reactions at the Alpha-Carbon1h 53m
- Tautomerization9m
- Tautomers of Dicarbonyl Compounds6m
- Enolate4m
- Acid-Catalyzed Alpha-Halogentation4m
- Base-Catalyzed Alpha-Halogentation3m
- Haloform Reaction8m
- Hell-Volhard-Zelinski Reaction3m
- Overview of Alpha-Alkylations and Acylations5m
- Enolate Alkylation and Acylation12m
- Enamine Alkylation and Acylation16m
- Beta-Dicarbonyl Synthesis Pathway7m
- Acetoacetic Ester Synthesis13m
- Malonic Ester Synthesis15m
- 25. Condensation Chemistry2h 9m
- 26. Amines1h 43m
- 27. Heterocycles2h 0m
- Nomenclature of Heterocycles15m
- Acid-Base Properties of Nitrogen Heterocycles10m
- Reactions of Pyrrole, Furan, and Thiophene13m
- Directing Effects in Substituted Pyrroles, Furans, and Thiophenes16m
- Addition Reactions of Furan8m
- EAS Reactions of Pyridine17m
- SNAr Reactions of Pyridine18m
- Side-Chain Reactions of Substituted Pyridines20m
- 28. Carbohydrates5h 53m
- Monosaccharide20m
- Monosaccharides - D and L Isomerism9m
- Monosaccharides - Drawing Fischer Projections18m
- Monosaccharides - Common Structures6m
- Monosaccharides - Forming Cyclic Hemiacetals12m
- Monosaccharides - Cyclization18m
- Monosaccharides - Haworth Projections13m
- Mutarotation11m
- Epimerization9m
- Monosaccharides - Aldose-Ketose Rearrangement8m
- Monosaccharides - Alkylation10m
- Monosaccharides - Acylation7m
- Glycoside6m
- Monosaccharides - N-Glycosides18m
- Monosaccharides - Reduction (Alditols)12m
- Monosaccharides - Weak Oxidation (Aldonic Acid)7m
- Reducing Sugars23m
- Monosaccharides - Strong Oxidation (Aldaric Acid)11m
- Monosaccharides - Oxidative Cleavage27m
- Monosaccharides - Osazones10m
- Monosaccharides - Kiliani-Fischer23m
- Monosaccharides - Wohl Degradation12m
- Monosaccharides - Ruff Degradation12m
- Disaccharide30m
- Polysaccharide11m
- 29. Amino Acids3h 20m
- Proteins and Amino Acids19m
- L and D Amino Acids14m
- Polar Amino Acids14m
- Amino Acid Chart18m
- Acid-Base Properties of Amino Acids33m
- Isoelectric Point14m
- Amino Acid Synthesis: HVZ Method12m
- Synthesis of Amino Acids: Acetamidomalonic Ester Synthesis16m
- Synthesis of Amino Acids: N-Phthalimidomalonic Ester Synthesis13m
- Synthesis of Amino Acids: Strecker Synthesis13m
- Reactions of Amino Acids: Esterification7m
- Reactions of Amino Acids: Acylation3m
- Reactions of Amino Acids: Hydrogenolysis6m
- Reactions of Amino Acids: Ninhydrin Test11m
- 30. Peptides and Proteins2h 42m
- Peptides12m
- Primary Protein Structure4m
- Secondary Protein Structure17m
- Tertiary Protein Structure11m
- Disulfide Bonds17m
- Quaternary Protein Structure10m
- Summary of Protein Structure7m
- Intro to Peptide Sequencing2m
- Peptide Sequencing: Partial Hydrolysis25m
- Peptide Sequencing: Partial Hydrolysis with Cyanogen Bromide7m
- Peptide Sequencing: Edman Degradation28m
- Merrifield Solid-Phase Peptide Synthesis18m
- 32. Lipids 2h 50m
- 34. Nucleic Acids1h 32m
- 35. Transition Metals5h 33m
- Electron Configuration of Elements45m
- Coordination Complexes20m
- Ligands24m
- Electron Counting10m
- The 18 and 16 Electron Rule13m
- Cross-Coupling General Reactions40m
- Heck Reaction40m
- Stille Reaction13m
- Suzuki Reaction25m
- Sonogashira Coupling Reaction17m
- Fukuyama Coupling Reaction15m
- Kumada Coupling Reaction13m
- Negishi Coupling Reaction16m
- Buchwald-Hartwig Amination Reaction19m
- Eglinton Reaction17m
Hydrolysis of Nucleosides - Online Tutor, Practice Problems & Exam Prep
 Created using AI
Created using AIThe hydrolysis of nucleosides, though not easily achieved, can occur under strong acidic conditions over time, yielding a free sugar and a nitrogenous base. The mechanism involves four key steps: proton transfer, loss of the leaving group, nucleophilic attack, and another proton transfer. Understanding these steps is crucial for grasping the overall process of acid-catalyzed hydrolysis, which is significant in biochemical reactions involving nucleotides and nucleic acids.
Hydrolysis of Nucleosides Concept 1
Video transcript
Hydrolysis of Nucleosides Example 1
Video transcript
Hey everyone. So in this example question, it says, provide the mechanism for the acid catalyzed hydrolysis of Caedidine. Step 1, we're going to use the hydronium ion, and we're going to use it to protonate the carbonyl oxygen. Here is our carbonyl oxygen. We're going to introduce our hydronium ion, which is H3O+. It's water mixed with acid here that we would do this. The HCl here is just helping provide an acidic enough environment that we can protonate the water. As a result, we create H3O+. So we're going to protonate this carbon and oxygen. Doing that gives us this structure initially. We still have this CH2OH portion here. We have our two OH groups. We have our nitrogenous base. And the nitrogenous base has its lone pair. Here goes our oxygen now, which is positively charged because it gained that H+.
Now, moving to step 2, we create a double bond between the anomeric carbon and the anomeric oxygen to expel the base. Remember, our anomeric oxygen is this oxygen right here, and the anomeric carbon is this carbon here. They're going to form a double bond with each other. Carbon can only make four bonds at one time, so that's going to expel this entire base here. Now we have this temporary structure. Here's our double bond between the anomeric carbon and the anomeric oxygen. We still have our two OH groups. We have our CH2OH group, plus our expelled base. The H from the bond now belongs to it. Here is our free sugar and our base.
For our nucleophilic attack, we're going to use water as a nucleophile to attack the anomeric carbon. Water is going to come in and hit this anomeric carbon, kicking this bond back to the oxygen. We still have our base just sitting there. Now, at this point, here's our sugar. We have our OH groups. We have our CH2OH. And now we have our oxygen, which is attached. This water, which detaches, can attach from the top or bottom, so either stereochemistry of a wedge or dash bond is possible. To show this, we show this squiggly line. Now, oxygen is making three bonds, so it's positively charged. And we still have this base here in its same form.
Now we use another water to deprotonate our positively charged oxygen. We can't just leave it like this; we can't have a charge on our final product. A second water molecule comes in. It's going to deprotonate, so it's going to remove this H. Oxygen holds on to the electrons. Here is our sugar again. The OH groups are still present. We have our CH2OH. Again, we don't know the stereochemistry of this OH group. It could align itself top or bottom and give us alpha or beta of the sugar. We're just showing the squiggly line to show both are possible. Here is our nitrogenous base. Technically, this nitrogenous base could be in this form, its enol form, or it could be in its keto form, where you have the nitrogens here. N and N. So we're showing the tautomers of this particular base. Either one is a possibility. But here we have our free sugar, and we have our base that has been formed. And here I've shown both tautomers of that particular base.
This will represent the acid catalyzed mechanism when it comes to breaking down this nucleoside yet again into its free sugar and its base.
Propose a mechanism for acid-catalyzed hydrolysis of adenosine.
Problem Transcript
Do you want more practice?
More setsHere’s what students ask on this topic:
What is the mechanism of acid-catalyzed hydrolysis of nucleosides?
The acid-catalyzed hydrolysis of nucleosides involves four key steps:
1. Proton Transfer: The nucleoside's glycosidic bond is protonated by the acid, making it more susceptible to cleavage.
2. Loss of Leaving Group: The protonated glycosidic bond breaks, releasing the nitrogenous base and forming a carbocation intermediate.
3. Nucleophilic Attack: Water acts as a nucleophile and attacks the carbocation, leading to the formation of a protonated sugar.
4. Proton Transfer: The protonated sugar loses a proton, resulting in the formation of a free sugar and completing the hydrolysis process.
This mechanism is crucial for understanding biochemical reactions involving nucleotides and nucleic acids.
 Created using AI
Created using AIWhat are the products of nucleoside hydrolysis?
The hydrolysis of nucleosides under strong acidic conditions yields two main products: a free sugar and a nitrogenous base. The free sugar is typically a ribose or deoxyribose, depending on whether the nucleoside is a ribonucleoside or a deoxyribonucleoside. The nitrogenous base can be one of the four bases found in nucleic acids: adenine, guanine, cytosine, or thymine (uracil in RNA). Understanding these products is essential for studying the biochemical pathways involving nucleotides and nucleic acids.
 Created using AI
Created using AIWhy are nucleosides not easily hydrolyzed?
Nucleosides are not easily hydrolyzed because the glycosidic bond between the sugar and the nitrogenous base is relatively stable under normal conditions. This bond requires strong acidic conditions and sufficient time to break. The stability of the glycosidic bond is crucial for the integrity of nucleic acids, ensuring that genetic information is preserved and not easily degraded. However, under strong acidic conditions, the bond can be protonated, making it more susceptible to cleavage and allowing hydrolysis to occur.
 Created using AI
Created using AIWhat role does proton transfer play in the hydrolysis of nucleosides?
Proton transfer plays a critical role in the hydrolysis of nucleosides. It occurs in two key steps of the mechanism:
1. Initial Protonation: The glycosidic bond is protonated by the acid, making it more susceptible to cleavage.
2. Final Deprotonation: After the nucleophilic attack by water, the protonated sugar loses a proton, resulting in the formation of a free sugar.
These proton transfer steps are essential for facilitating the cleavage of the glycosidic bond and completing the hydrolysis process.
 Created using AI
Created using AIHow does the hydrolysis of nucleosides differ from the hydrolysis of nucleotides?
The hydrolysis of nucleosides and nucleotides involves different processes and products:
Nucleosides: Hydrolysis of nucleosides under strong acidic conditions yields a free sugar and a nitrogenous base. The mechanism involves proton transfer, loss of the leaving group, nucleophilic attack, and another proton transfer.
Nucleotides: Hydrolysis of nucleotides involves breaking the phosphodiester bond, resulting in the release of a phosphate group and a nucleoside. This process can occur under both acidic and basic conditions and is crucial for various biochemical pathways, including DNA and RNA degradation.
Understanding these differences is important for studying the biochemical roles of nucleosides and nucleotides.
 Created using AI
Created using AI