This video, we're going to begin our lesson on DNA fingerprinting. DNA fingerprinting is really just a specific technique that is going to use genetic markers within a genome to identify an individual. Just like a fingerprint can be used to identify an individual, DNA fingerprinting can help to identify an individual. What are these genetic markers? Genetic markers are sequences of DNA with a known location and are easily identifiable in a genome. Markers specifically refer to polymorphisms, and polymorphisms are completely different between the genomes of each individual, so they're differences in the sequence across different individuals. For example, single nucleotide polymorphisms, which are commonly abbreviated as SNPs, are genetic markers in a genome that differ by just one single nucleotide across different organisms. If we take a look at an example image, we're looking at how a single nucleotide polymorphism can exist between 2 alleles within the same organism across 2 different individuals. In this image below, what you'll notice is that we have individual number 1, which is right here, and this is the specific DNA sequence of individual number 1. Below, we have individual number 2, whose sequence is right here. And what you'll notice is that the DNA sequence of individual 1 and individual number 2 are pretty much exactly the same, except for one position of this base pair. This would be referred to as a single nucleotide polymorphism or a SNP because it's just this one position here that is going to differ between the two, where individual one has a TA base pair at this position, whereas individual 2 has a CG base pair. These polymorphisms are going to be unique to specific individuals and can be used to help identify an individual just like a fingerprint can be used to identify an individual. Now, a person's DNA fingerprint is really just referring to the combination of all the unique genetic markers in an individual's genome. This here concludes our brief introduction to DNA fingerprinting and how these unique genetic markers and polymorphisms can be used to help identify an individual. We'll be able to get some practice applying these concepts as we move forward. So I'll see you all in our next video.
- 1. Introduction to Microbiology3h 21m
- Introduction to Microbiology16m
- Introduction to Taxonomy26m
- Scientific Naming of Organisms9m
- Members of the Bacterial World10m
- Introduction to Bacteria9m
- Introduction to Archaea10m
- Introduction to Eukarya20m
- Acellular Infectious Agents: Viruses, Viroids & Prions19m
- Importance of Microorganisms20m
- Scientific Method27m
- Experimental Design30m
- 2. Disproving Spontaneous Generation1h 18m
- 3. Chemical Principles of Microbiology3h 38m
- 4. Water1h 28m
- 5. Molecules of Microbiology2h 23m
- 6. Cell Membrane & Transport3h 28m
- Cell Envelope & Biological Membranes12m
- Bacterial & Eukaryotic Cell Membranes8m
- Archaeal Cell Membranes18m
- Types of Membrane Proteins8m
- Concentration Gradients and Diffusion9m
- Introduction to Membrane Transport14m
- Passive vs. Active Transport13m
- Osmosis33m
- Simple and Facilitated Diffusion17m
- Active Transport30m
- ABC Transporters11m
- Group Translocation7m
- Types of Small Molecule Transport Review9m
- Endocytosis and Exocytosis15m
- 7. Prokaryotic Cell Structures & Functions5h 52m
- Prokaryotic & Eukaryotic Cells26m
- Binary Fission11m
- Generation Times16m
- Bacterial Cell Morphology & Arrangements35m
- Overview of Prokaryotic Cell Structure10m
- Introduction to Bacterial Cell Walls26m
- Gram-Positive Cell Walls11m
- Gram-Negative Cell Walls20m
- Gram-Positive vs. Gram-Negative Cell Walls11m
- The Glycocalyx: Capsules & Slime Layers12m
- Introduction to Biofilms6m
- Pili18m
- Fimbriae & Hami7m
- Introduction to Prokaryotic Flagella12m
- Prokaryotic Flagellar Structure18m
- Prokaryotic Flagellar Movement11m
- Proton Motive Force Drives Flagellar Motility5m
- Chemotaxis14m
- Review of Prokaryotic Surface Structures8m
- Prokaryotic Ribosomes16m
- Introduction to Bacterial Plasmids13m
- Cell Inclusions9m
- Endospores16m
- Sporulation5m
- Germination5m
- 8. Eukaryotic Cell Structures & Functions2h 18m
- 9. Microscopes2h 46m
- Introduction to Microscopes8m
- Magnification, Resolution, & Contrast10m
- Introduction to Light Microscopy5m
- Light Microscopy: Bright-Field Microscopes23m
- Light Microscopes that Increase Contrast16m
- Light Microscopes that Detect Fluorescence16m
- Electron Microscopes14m
- Reviewing the Different Types of Microscopes10m
- Introduction to Staining5m
- Simple Staining14m
- Differential Staining6m
- Other Types of Staining11m
- Reviewing the Types of Staining8m
- Gram Stain13m
- 10. Dynamics of Microbial Growth4h 36m
- Biofilms16m
- Growing a Pure Culture5m
- Microbial Growth Curves in a Closed System21m
- Temperature Requirements for Microbial Growth18m
- Oxygen Requirements for Microbial Growth22m
- pH Requirements for Microbial Growth8m
- Osmolarity Factors for Microbial Growth14m
- Reviewing the Environmental Factors of Microbial Growth12m
- Nutritional Factors of Microbial Growth30m
- Growth Factors4m
- Introduction to Cultivating Microbial Growth5m
- Types of Solid Culture Media4m
- Plating Methods16m
- Measuring Growth by Direct Cell Counts9m
- Measuring Growth by Plate Counts14m
- Measuring Growth by Membrane Filtration6m
- Measuring Growth by Biomass15m
- Introduction to the Types of Culture Media5m
- Chemically Defined Media3m
- Complex Media4m
- Selective Media5m
- Differential Media9m
- Reducing Media4m
- Enrichment Media7m
- Reviewing the Types of Culture Media8m
- 11. Controlling Microbial Growth4h 10m
- Introduction to Controlling Microbial Growth29m
- Selecting a Method to Control Microbial Growth44m
- Physical Methods to Control Microbial Growth49m
- Review of Physical Methods to Control Microbial Growth7m
- Chemical Methods to Control Microbial Growth16m
- Chemicals Used to Control Microbial Growth6m
- Liquid Chemicals: Alcohols, Aldehydes, & Biguanides15m
- Liquid Chemicals: Halogens12m
- Liquid Chemicals: Surface-Active Agents17m
- Other Types of Liquid Chemicals14m
- Chemical Gases: Ethylene Oxide, Ozone, & Formaldehyde13m
- Review of Chemicals Used to Control Microbial Growth11m
- Chemical Preservation of Perishable Products10m
- 12. Microbial Metabolism5h 16m
- Introduction to Energy15m
- Laws of Thermodynamics15m
- Chemical Reactions9m
- ATP20m
- Enzymes14m
- Enzyme Activation Energy9m
- Enzyme Binding Factors9m
- Enzyme Inhibition10m
- Introduction to Metabolism8m
- Negative & Positive Feedback7m
- Redox Reactions22m
- Introduction to Aerobic Cellular Respiration25m
- Types of Phosphorylation12m
- Glycolysis19m
- Entner-Doudoroff Pathway11m
- Pentose-Phosphate Pathway10m
- Pyruvate Oxidation8m
- Krebs Cycle16m
- Electron Transport Chain19m
- Chemiosmosis7m
- Review of Aerobic Cellular Respiration19m
- Fermentation & Anaerobic Respiration23m
- 13. Photosynthesis2h 31m
- 14. DNA Replication2h 25m
- 15. Central Dogma & Gene Regulation7h 14m
- Central Dogma7m
- Introduction to Transcription20m
- Steps of Transcription22m
- Transcription Termination in Prokaryotes7m
- Eukaryotic RNA Processing and Splicing20m
- Introduction to Types of RNA9m
- Genetic Code25m
- Introduction to Translation30m
- Steps of Translation23m
- Review of Transcription vs. Translation12m
- Prokaryotic Gene Expression21m
- Review of Prokaryotic vs. Eukaryotic Gene Expression13m
- Introduction to Regulation of Gene Expression13m
- Prokaryotic Gene Regulation via Operons27m
- The Lac Operon21m
- Glucose's Impact on Lac Operon25m
- The Trp Operon20m
- Review of the Lac Operon & Trp Operon11m
- Introduction to Eukaryotic Gene Regulation9m
- Eukaryotic Chromatin Modifications16m
- Eukaryotic Transcriptional Control22m
- Eukaryotic Post-Transcriptional Regulation28m
- Post-Translational Modification6m
- Eukaryotic Post-Translational Regulation13m
- 16. Microbial Genetics4h 44m
- Introduction to Microbial Genetics11m
- Introduction to Mutations20m
- Methods of Inducing Mutations15m
- Prototrophs vs. Auxotrophs13m
- Mutant Detection25m
- The Ames Test14m
- Introduction to DNA Repair5m
- DNA Repair Mechanisms37m
- Horizontal Gene Transfer18m
- Bacterial Transformation11m
- Transduction32m
- Introduction to Conjugation6m
- Conjugation: F Plasmids18m
- Conjugation: Hfr & F' Cells19m
- Genome Variability21m
- CRISPR CAS11m
- 17. Biotechnology3h 0m
- 18. Viruses, Viroids, & Prions4h 56m
- Introduction to Viruses20m
- Introduction to Bacteriophage Infections14m
- Bacteriophage: Lytic Phage Infections12m
- Bacteriophage: Lysogenic Phage Infections17m
- Bacteriophage: Filamentous Phage Infections8m
- Plaque Assays9m
- Introduction to Animal Virus Infections10m
- Animal Viruses: 1. Attachment to the Host Cell7m
- Animal Viruses: 2. Entry & Uncoating in the Host Cell19m
- Animal Viruses: 3. Synthesis & Replication22m
- Animal Viruses: DNA Virus Synthesis & Replication14m
- Animal Viruses: RNA Virus Synthesis & Replication22m
- Animal Viruses: Antigenic Drift vs. Antigenic Shift9m
- Animal Viruses: Reverse-Transcribing Virus Synthesis & Replication9m
- Animal Viruses: 4. Assembly Inside Host Cell8m
- Animal Viruses: 5. Release from Host Cell15m
- Acute vs. Persistent Viral Infections25m
- COVID-19 (SARS-CoV-2)14m
- Plant Viruses12m
- Viroids6m
- Prions13m
- 19. Innate Immunity7h 15m
- Introduction to Immunity8m
- Introduction to Innate Immunity17m
- Introduction to First-Line Defenses5m
- Physical Barriers in First-Line Defenses: Skin13m
- Physical Barriers in First-Line Defenses: Mucous Membrane9m
- First-Line Defenses: Chemical Barriers24m
- First-Line Defenses: Normal Microflora5m
- Introduction to Cells of the Immune System15m
- Cells of the Immune System: Granulocytes29m
- Cells of the Immune System: Agranulocytes25m
- Introduction to Cell Communication5m
- Cell Communication: Surface Receptors & Adhesion Molecules16m
- Cell Communication: Cytokines27m
- Pattern Recognition Receptors (PRRs)45m
- Introduction to the Complement System24m
- Activation Pathways of the Complement System23m
- Effects of the Complement System23m
- Review of the Complement System12m
- Phagoctytosis21m
- Introduction to Inflammation18m
- Steps of the Inflammatory Response26m
- Fever8m
- Interferon Response25m
- 20. Adaptive Immunity7h 14m
- Introduction to Adaptive Immunity32m
- Antigens12m
- Introduction to T Lymphocytes38m
- Major Histocompatibility Complex Molecules20m
- Activation of T Lymphocytes21m
- Functions of T Lymphocytes25m
- Review of Cytotoxic vs Helper T Cells13m
- Introduction to B Lymphocytes27m
- Antibodies14m
- Classes of Antibodies35m
- Outcomes of Antibody Binding to Antigen15m
- T Dependent & T Independent Antigens21m
- Clonal Selection20m
- Antibody Class Switching17m
- Affinity Maturation14m
- Primary and Secondary Response of Adaptive Immunity21m
- Immune Tolerance28m
- Regulatory T Cells10m
- Natural Killer Cells16m
- Review of Adaptive Immunity25m
- 21. Principles of Disease6h 57m
- Symbiotic Relationships12m
- The Human Microbiome46m
- Characteristics of Infectious Disease47m
- Stages of Infectious Disease Progression26m
- Koch's Postulates26m
- Molecular Koch's Postulates11m
- Bacterial Pathogenesis36m
- Introduction to Pathogenic Toxins6m
- Exotoxins Cause Damage to the Host40m
- Endotoxin Causes Damage to the Host13m
- Exotoxins vs. Endotoxin Review13m
- Immune Response Damage to the Host15m
- Introduction to Avoiding Host Defense Mechanisms8m
- 1) Hide Within Host Cells5m
- 2) Avoiding Phagocytosis31m
- 3) Surviving Inside Phagocytic Cells10m
- 4) Avoiding Complement System9m
- 5) Avoiding Antibodies25m
- Viruses Evade the Immune Response27m
DNA Fingerprinting - Online Tutor, Practice Problems & Exam Prep
 Created using AI
Created using AIDNA fingerprinting utilizes genetic markers, specifically polymorphisms like single nucleotide polymorphisms (SNPs), to identify individuals uniquely. These markers differ among individuals, similar to traditional fingerprints. Short tandem repeats (STRs), which are 2 to 5 nucleotide-long sequences, also serve as genetic markers, varying in number across individuals. Both SNPs and STRs are crucial in forensic science for identifying suspects through DNA evidence found at crime scenes, highlighting their importance in genetic identification and criminal investigations.
DNA Fingerprinting
Video transcript
Short Tandem Repeats (STRs)
Video transcript
In this video, we're going to introduce short tandem repeats, which are commonly abbreviated as STRs. These genetic markers used by researchers are generally made up of short repeat sequences that vary in number. These repeats are referred to as short tandem repeats, or STRs, which are short repeated sequences of DNA that are approximately 2 to 5 nucleotides long. Nucleotides, abbreviated as Nts, are found in very specific regions of a genome. The specific number of STRs in this region of a genome is polymorphic, meaning that it is unique for each person and can be used to identify individuals. Here in this example image below, we are showing you some examples of short tandem repeats. Short tandem repeats, or STRs, are genetic markers that can be used to identify individuals, perhaps by using DNA found at a crime scene. These markers can help solve a crime.
Let's take a look at the image below. Notice that the repeated sequence, the short tandem repeat we are focusing on is the sequence 'g a t a'. This is the double-stranded DNA and the specific short tandem repeat in focus. You will notice DNA from three different individuals below. Individual one's DNA is here; then we have individual two, their DNA is there, and individual three's DNA is at the bottom. In this specific region of interest in the chromosome, each of these three individuals differs in their number of short tandem repeats or STRs. For instance, individual number one has a total of 5 STRs, which is unique to this specific individual. Individual number two has only 3 of these short tandem repeats within this region of the genome, which is unique to them. Individual number three has a total of 4 short tandem repeats. The number of short tandem repeats is unique for each individual and serves as a genetic marker that can be used to identify an individual and help solve a crime by comparing DNA that might be found at a crime scene with the DNA from suspects.
This concludes our brief introduction to short tandem repeats or STRs, and we'll be able to get some practice applying these concepts as we move forward in our course. So, I'll see you all in our next video.
The goal of DNA fingerprinting is:
Which of the following characteristics of short tandem repeats (STRs) makes it useful for DNA fingerprinting?
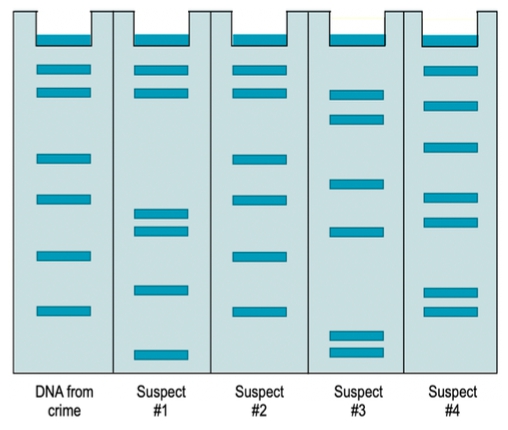
The gel below shows a region of STRs from a DNA sample taken from a crime scene. It also shows the same region of STRs from 4 suspects involved in the case. Which suspect' DNA was found at the crime scene?
Do you want more practice?
Here’s what students ask on this topic:
What is DNA fingerprinting and how is it used to identify individuals?
DNA fingerprinting is a technique that uses genetic markers within a genome to uniquely identify an individual. These genetic markers, such as single nucleotide polymorphisms (SNPs) and short tandem repeats (STRs), vary between individuals. SNPs are differences in a single nucleotide, while STRs are short sequences of DNA that repeat a variable number of times. By analyzing these polymorphisms, scientists can create a unique DNA profile for each person. This method is widely used in forensic science to match DNA from crime scenes with suspects, as well as in paternity testing and genetic research.
 Created using AI
Created using AIWhat are single nucleotide polymorphisms (SNPs) and how do they contribute to DNA fingerprinting?
Single nucleotide polymorphisms (SNPs) are genetic markers that differ by a single nucleotide between individuals. For example, at a specific position in the DNA sequence, one person might have an adenine (A) while another has a cytosine (C). These variations are unique to each individual and can be used to create a DNA fingerprint. In DNA fingerprinting, multiple SNPs are analyzed to generate a unique genetic profile, which can be used for identification purposes in forensic science, paternity testing, and genetic research.
 Created using AI
Created using AIWhat are short tandem repeats (STRs) and why are they important in forensic science?
Short tandem repeats (STRs) are short sequences of DNA, typically 2 to 5 nucleotides long, that repeat multiple times in a row. The number of these repeats varies between individuals, making STRs useful genetic markers for identification. In forensic science, STR analysis is used to compare DNA samples from crime scenes with those of suspects. By examining the number of repeats at specific STR loci, forensic scientists can create a DNA profile that is unique to each individual, aiding in criminal investigations and legal proceedings.
 Created using AI
Created using AIHow do genetic markers like SNPs and STRs differ in their application in DNA fingerprinting?
Both SNPs and STRs are used as genetic markers in DNA fingerprinting, but they differ in their characteristics and applications. SNPs involve a single nucleotide difference in the DNA sequence, making them highly specific and useful for fine-scale genetic analysis. STRs, on the other hand, consist of short sequences that repeat a variable number of times, providing a broader range of variation. While SNPs are often used for detailed genetic studies and ancestry testing, STRs are more commonly used in forensic science for creating DNA profiles due to their high variability and ease of analysis.
 Created using AI
Created using AIHow is DNA fingerprinting used in criminal investigations?
In criminal investigations, DNA fingerprinting is used to match DNA evidence from a crime scene with potential suspects. By analyzing genetic markers such as SNPs and STRs, forensic scientists can create a DNA profile from the evidence. This profile is then compared to the DNA profiles of suspects. If a match is found, it can provide strong evidence linking a suspect to the crime scene. DNA fingerprinting is a powerful tool in solving crimes, identifying victims, and exonerating innocent individuals.
 Created using AI
Created using AI