Again, it's a little out of order, but I'm actually going to talk about complementation first. So, a complementation test is performed in order to determine if two mutants with the same phenotype have mutations in the same gene. So if you do this huge, like, cross or this huge experiment with flies, for instance, and you have thousands of them, and you come across a couple with the same gene or the same phenotype. They all have really short legs, for instance. So you can do a complementation test where you mate them together to figure out whether or not those short legs mutations that you find are all in the same gene, like a short leg gene, or if there are mutations in multiple genes causing the same phenotype. So how you perform a complementation test is you take the two mutants that you have, and if they're recessive mutations, you can mate them together. And when you do that, if the offspring are wild type, meaning that they have long legs, they don't have that short leg phenotype, then the two mutations are in different genes. If they're mutant, that means the two mutations are in the same genes. Same genes, not different, same. Now, sorry about that. I will go back and edit that so it's clear in your handout. But just know, these are in the same genes. Now, why is that the case? Well, because if you have two mutants, say, mutant one and mutant two. And, here's their mutations. If they're in the same genes, when you do this cross, you're going to get them all with mutants. Right? That's what the cross is going to look like. But if instead, it's in two genes, so what this would look like is this, you would have rr and ss, and ss. Right? So these the ones with the pluses are the wild type, the without or without. Then you would do a dihybrid cross. Because now you're looking at two different genes. Now, I'm not going to fill out. But just know that you're not going to get this recessive phenotype with a dihybrid cross because all of the offspring will have a wild type of both genes. So they all appear wild type. So that's how that works. So let's look at an example here. So say you have three white mutants, one, two, and three, and you want to know if the mutation causing them to be white is in the same gene for each mutant, and the wild type color is normally blue. So this question is asking which mutations complement. And so what if you get the question like that, what does that mean? It says, which mutations complement, meaning that the two mutations from the two organisms are in different genes. So they complement, not in the same gene, they're in different genes. So here you have, you're doing three crosses. Right? You're doing white one with white two, white one with white three, and white two with white three. And therefore, that gives you all the mating possibilities that you could do with these three mutants. And you can see here that these all result in different things. Some of them are white, and two of them are blue. So which mutations here complement, meaning that the mutations are in different genes? Well, let's go back up here to the rules and find out. If the mutations are wild type, which in this case is going to be blue, then the two mutations are in different genes. So, here we have this one, and here we have this one. So white one and three and white two and three complement. And usually, it's going to be you're going to see white three because that's the common factor. So white three complements twelve. It's mostly how you're going to see that. But if one is all white, if the mutations are all mutant, which in this case, in this problem is white, then the two mutations are in the same gene. And so this is not complement because they're in the same gene. These two are in different genes. So that's how you do a complementation test. You're definitely going to be asked about this. But just remember here, if the offspring have the wild type phenotype, they're in different genes, meaning that they complement. If their offspring are all mutant, they're in the same gene, means they don't complement. So with that, let's now move on.
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
Epistasis and Complementation - Online Tutor, Practice Problems & Exam Prep
 Created using AI
Created using AIComplementation tests determine if mutations causing the same phenotype are in the same gene. If offspring from two mutants show a wild type phenotype, the mutations are in different genes; if all offspring are mutant, they are in the same gene. Gene interactions include nonepistatic genes, which do not mask each other's effects, and epistatic genes, where one gene's dominant allele masks another's phenotype. Dominant epistasis results in a 12:3:1 ratio, while recessive epistasis yields a 9:3:4 ratio. Understanding these interactions is crucial for genetic analysis.
Complementation
Video transcript
Non-Epistatic Genes
Video transcript
Okay, so now let's talk about nonepistatic genes, and these are going to be genes that, they're two genes, typically two or more genes, and, they affect the same phenotype, but they're not necessarily interacting. This is different from epistasis, which is the interaction of two genes. Now I'm presenting nonepistatic genes first because this is going to be the closest to what you're familiar with, and then we can base everything else, all the epistatic genes, in comparison to this. So let me back up here. First, epistasis, and generally when this happens, the presence of one gene allele will mask the phenotype of the second gene allele. So remember, we're talking about nonepistatic here, so nonepistatic, they won't mask it. Epistatic, they will. And so in non-epistatic situations, you're going to see the same ratio that you're used to seeing, the 9:3:3:1. In the epistatic, you're going to see an altered ratio. It’s going to be different, and different types of epistasis have different ratios, and we'll go through all of these individually. But for now, let's talk about this example. This is going to be focusing on corn snakes. Corn snakes come in four colors: orange, black, camouflage, and albino. Here's an example of a corn snake. Here, you can see that it has orange and black, so this would be a camouflage coloration. There are two genes that control this, O and B. Obviously, O is for orange, B is for black. Now, there are many different genotypes you can have, and you're going to see these charts a lot more coming up, so I want to explain it now. So here you can see that you have a dominant here and a line here. So what does the line mean? The line means it can either be dominant or recessive. So here we have homozygous or heterozygous dominant for O and homozygous recessive for B. So in this genotype, you're going to get orange. In the opposite, meaning that this is homozygous recessive for O and homo or heterozygous dominant for B, you're going to get black. Now if you have a dominant allele for both O and B, you're going to get the camouflage, which is what this looks like where you can see both O and B, or the orange and the black, and if you're recessive for both, you're going to get albino.
So, we know this about this thing. I'm giving this to you. So we do a question like this. It says, What is the offspring's genotype and phenotype derived from the mating of two heterozygous camouflage corn snakes? So, heterozygous camouflage is going to be heterozygous for both O and B, and because it's a mating, there are two of them. So, you can do a Punnett square if you prefer, but I think that the branch method is going to be faster. You want to do a Punnett square, feel free to pause it right now. Write out your Punnett square, or even pause it if you're going to do the branch method. See if you can figure out what the genotypes are, and the ratios for each, and then, come back and I'll do the explanation. So hopefully, you have paused it and now you're restarting it, and you're looking at the, to check your answer. So, we're going to walk through. So, we're doing this heterozygous mating with, orange and black. It's camouflaged here. If we're going to start with this, our first is orange or not orange. So, how do we do this? We do a basic Punnett Square. So, 3/4 will be orange, and 1/4 will be not orange, because the two recessive o's here can be a bunch of different colors. It can be albino, it can be black, but just for right now, we're saying not orange. Then, for each, we want to do the same for black. Now, I can write out the Punnett square, but it's going to look like this,916×916. 316×116. So, what are the offspring's genotype and phenotype? Well, 916 will be camouflage, which means heterozygous for both. 316 will be orange. 316 will be black, and 116 will be albino. And so when we see this 9:3:3:1 ratio in a situation in which, you know, two genes are working together to interact with the same phenotype, this is going to be a nonepistatic situation. So, this is the normal ratio that you're used to seeing, so this is the nonepistatic situation. So now, let's move on and get to epistatic situations.
Epistatic Genes
Video transcript
Okay. So now we're going to talk about epistatic genes. This is going to be a long video, so you're going to have to stick with me. The reason is that I'm going over dominant epistasis and recessive epistasis, which are different and have different phenotypic ratios at the end. So just bear with me, and I'm going to give you some really great examples of both of these cases that you will need to know because you're going to see them.
The important thing about this is that with each type of epistasis and everything that I present from now on, I'm going to be giving you a ratio. It's not going to be 9 to 3 to 3 to 1, but it's going to be something different. And you'll need to know what ratio goes with what topic. So, be sure that you're writing these ratios down because you will see these ratios in a test situation, and you're going to have to know which one goes with which.
The first thing that I'm talking about is dominant epistasis. This occurs when a dominant allele of one gene is masking the effect of a second gene. Remember, epistasis is talking about two gene interactions. There are two genes here; one of them is dominant, and because that dominant allele is present, it covers up or masks the phenotype of the other allele, whether or not it's dominant or recessive. Here, we say that the dominant allele is epistatic, and the phenotypic ratio of a cross from a heterozygous cross is not 9 to 3 to 3 to 1 but instead 12 to 3 to 1. Memorize this and associate it with dominant epistasis because 12 to 3 to 1 always means dominant epistasis.
We're dealing with a certain breed of squash that comes in 3 colors: white, dark red, and light red. Coloration is determined by two genes, d and w. For example, if you have dominant d and dominant w, it doesn't matter whether they are homozygous dominant or heterozygous. If there’s at least one dominant allele, you get a white phenotype. If you are recessive for the d and dominant for the w, you also get a white phenotype. However, if you have a dominant d and recessive w, you get a dark red, and if recessive for both, you get light red. Here, we say the dominant w allele is epistatic because, anytime it's present, it determines the phenotype you will see.
Therefore, in the presence of dominant w, the phenotype will always be white, regardless of the other allele’s state. This information leads us to the phenotypic ratio of 12 to 3 to 1 because both cases involving dominant w results in white. The genotypic ratio remains 9 to 3 to 3 to 1, which can be confirmed via a dihybrid Punnett square or a branch diagram.
However, there is an alternate form called recessive epistasis, where the recessive allele masks the phenotype of the second gene with a different ratio of 9 to 3 to 4. For example, consider a breed of flower with colors of blue, magenta, and white, determined by two genes. If both genes are wild type, you get blue; if wild type for w but mutant for m, you get magenta; if mutant for w and wild type for m, you get white; and if mutant for both, you also get white. Here, the mutant w allele is epistatic and recessive because it must be present in two copies to affect the phenotype.
This phenomenon also has real-life implications, such as the Bombay Phenotype in humans, which involves blood types and is a type of recessive epistasis. This deals with the I and h gene families where the rare h mutation masks the expression of any I allele leading to a perceivable blood type O regardless of the I genotypes present. Thus, recessive epistasis can play significant roles beyond academic tests or breeding studies, highlighting its importance in fields like genetics.
Understanding these concepts, their phenotypic and genotypic ratios, and being able to distinguish them is vital for anyone studying genetics. It’s important to know the differences between dominant and recessive epistasis, their implications, and their applications, which can be decisive in various biological and genetic contexts.
Other Gene Interactions
Video transcript
Okay. So now, let's talk about other types of gene interactions. The first one I want to mention is the complementary gene action. This is when two genes interact because they are in the same pathway. So you have a single pathway that includes, you know, six or seven dozen genes, that produce proteins that interact in this pathway. Well, if you need one to start the other, this is complementary union action. The ratio here that you need to definitely know is 9 to 7. That's the super important one.
Let's look at an example here. A breed of flower comes in two colors, purple and white. Coloration is determined by two genes, c and p. So here we have, if you have a dominant c and a dominant p, you're going to get purple. If you have a dominant c but a recessive p, you'll get white. The same in the reverse with the recessive c and the dominant p, you'll get white, and recessive for both, you'll also get white. So, you have to be dominant in both the c and the p in order to get this color. The genotypic ratio will be 9 to 3 to 3 to 1. Don't believe me, do a Punnett square. But the phenotypic ratio will be 9 to 7 because these add up to seven and they're all white.
So, we say that these genes are complementary, and we're working through complementary gene action because both genes need to have a dominant allele in order to have that being a purple phenotype. This is complementary gene action.
There's another type, the second type called Suppressors, and these are mutant alleles. So now we're dealing with mutants, that dominance mutants. And the mutant of one gene will actually reverse the effect of a mutation in a second gene. So, now we're working with two mutations. There are two phenotypes that you can get here. The first, and this is the most common, the one that I'm going to present to you, and the one that you're most likely to be tested on is this. And, this is when the suppressor causes the phenotype to be like wild type. And it'll have a 13 to 3 ratio. The second, I'm not giving you an example of. It does exist. A couple of your books mention it, not all of you will even hear about it. But essentially, it's different because the suppressor causes the phenotype to be mutant, it has a different ratio. Feel free just to throw that into your memory just in case you're asked about it. But most of the time, if you're asked about a suppressor, it's going to be the 13 to 3 case.
So, an example of this is a breed of flower comes in two colors, the wild type red and the mutant purple. Coloration is determined by two genes, p and r. So, what we're dealing with here is if you have the wild type p and the dominant r, you get red. If you have the wild type p and the recessive r, you also get red because the wild type p is making it red. If you have the recessive p and the dominant r, you get purple. And the recessive in both, you get red. So in this case, the wild type p allele and the dominant r allele both are causing the plant to be red. So the only time that you get it to be purple is the mutant p allele. And this is because the recessive suppressor, which in this case is here, is suppressing the purple phenotype. Suppresses the purple phenotype. And so, that's why it's red. Because normally these both would cause it to be purple, but because this is a suppressor, it says, no, you're not going to be purple. I'm repressing you, and therefore, I'm going to be red, turns the plant red. Now, the genotypic ratio, again, is 9 to 3 to 3 to 1. However, the phenotypic ratio is going to be 13 to 3 because you have red, red, and red, and you have 3 purple. So, there's where that ratio comes from.
So, that's super important for a suppressor. And then, finally, the very or no, we have two more. So, this is a little different, though. These are modifiers, and this is when a mutation in one gene changes the degree of expression, so kind of how much it's expressed, of a mutated second gene. So here we go. So if you have, wild type at both genes, it's going to be wild type. If you have wild type at one and mutant at another, it's going to be defective in some way. So an example of this is it's defective. It has low transcription. If you have mutant at one and at the other, it's going to be also be defective, but in a different way. So, So now you have this mutated protein, and it does something different that doesn't have anything to do with transcription, but is a different pathway. And if you're mutated in both, they're extremely defective. So this is a modifier because the mutation at one gene affects the degree of expression of a mutated second gene. So these are modifying each other and, causing the, degree of expression to be defective or extremely defective. And, these are sort of a rare case. You many of you may not even be asked about these, but I wanted to throw it in there just in case that you were.
And then finally, synthetic lethals, and this is, lethal means dead, of course. So this is when two viable single mutations result in death when found as a double. So I'm not even giving you an example here, but here's just two dominants that are going to be purple, one dominant will be cyan, the other dominant will be white. But if you have, mutations in both, it's dead. So the genotypic ratio will be 9 to 3 to 3 to 1, but the phenotypic will be 9 to 3 to 3 because this one you won't see because it's dead. So, this is a unique case too. It's you may be asked about it, you may not. But just in case, I would memorize the ratios because that's how you're going to tell all of these different things apart. So if you see a 9 to 3 to 3 ratio, you know that this is because one of the alleles is a synthetic lethal. Meaning that when you have double mutants, so when you have mutated in this gene, the c gene, and the p gene, that causes it to be dead.
So, with that, let's now move on.
When performing a complementation test, how do you know if two mutations complement?
How can you tell if two genes are epistatic?
Two heterozygous organisms are crossed, and the F2 phenotypic ratio is 12:3:1. What form of epistasis do these two genes exhibit?
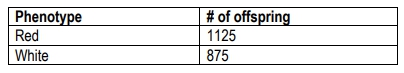
A cross of white plants and red plants was performed. Using the F2 phenotypic ratio data below, determine what form of gene interaction is taking place.
In the rare Bombay phenotype, a mutation in a second gene can control an individual's what?
Do you want more practice?
More setsHere’s what students ask on this topic:
What is a complementation test in genetics?
A complementation test is used to determine if two mutants with the same phenotype have mutations in the same gene. To perform this test, you cross the two mutants. If the offspring exhibit a wild type phenotype, the mutations are in different genes, indicating complementation. If the offspring are all mutant, the mutations are in the same gene, meaning no complementation. This test helps identify whether the observed phenotype is due to mutations in the same or different genes, which is crucial for understanding genetic pathways and gene interactions.
 Created using AI
Created using AIWhat is the difference between epistasis and complementation?
Epistasis refers to the interaction between two genes where one gene's allele masks the effect of another gene's allele. For example, in dominant epistasis, a dominant allele of one gene can mask the phenotype of another gene, resulting in a 12:3:1 phenotypic ratio. Complementation, on the other hand, is a test to determine if two mutants with the same phenotype have mutations in the same gene. If the offspring from the cross of two mutants show a wild type phenotype, the mutations are in different genes, indicating complementation.
 Created using AI
Created using AIWhat are the phenotypic ratios for dominant and recessive epistasis?
In dominant epistasis, the phenotypic ratio is 12:3:1. This occurs when a dominant allele of one gene masks the effect of another gene. In recessive epistasis, the phenotypic ratio is 9:3:4. This happens when the recessive allele of one gene masks the phenotype of another gene. These ratios are crucial for identifying the type of epistatic interaction in genetic studies.
 Created using AI
Created using AIHow do you interpret a 9:7 phenotypic ratio in genetics?
A 9:7 phenotypic ratio indicates complementary gene action. This occurs when two genes interact within the same pathway, and both need to have at least one dominant allele to produce a specific phenotype. For example, in a flower species, if both genes C and P need to be dominant to produce a purple color, any other combination (recessive in either or both genes) will result in a white color. This ratio helps identify genes that work together in a single pathway to produce a phenotype.
 Created using AI
Created using AIWhat is the Bombay phenotype in humans?
The Bombay phenotype is an example of recessive epistasis in humans, affecting blood type. Individuals with the Bombay phenotype have a rare mutation in the H gene, which prevents the addition of A or B antigens to the surface of red blood cells. As a result, even if they have the alleles for A or B blood types, they appear to have type O blood. This occurs because the H gene product is necessary for the expression of A and B antigens, and its absence masks the phenotype of the ABO blood group alleles.
 Created using AI
Created using AIYour Genetics tutor
- How did geneticists determine that inheritance of some phenotypic characteristics involves the interactions of...
- Define and distinguish epistasis and pleiotropy.
- A researcher interested in studying a human gene on chromosome 21 and another gene on the X chromosome uses FI...
- A researcher interested in studying a human gene on chromosome 21 and another gene on the X chromosome uses FI...
- In what way does position effect variegation (PEV) of Drosophila eye color indicate that chromatin state can a...
- Strains of petunias come in four pure-breeding colors: white, blue, red, and purple. White petunias are produc...
- Strains of petunias come in four pure-breeding colors: white, blue, red, and purple. White petunias are produc...
- Feather color in parakeets is produced by the blending of pigments from two biosynthetic pathways shown below....
- Feather color in parakeets is produced by the blending of pigments from two biosynthetic pathways shown below....
- Feather color in parakeets is produced by the blending of pigments from two biosynthetic pathways shown below....
- A male and a female mouse are each from pure-breeding albino strains. They have a litter of 10 pups, all of wh...
- A male and a female mouse are each from pure-breeding albino strains. They have a litter of 10 pups, all of wh...
- Three strains of green-seeded lentil plants appear to have the same phenotype. The strains are designated G₁, ...
- Three strains of green-seeded lentil plants appear to have the same phenotype. The strains are designated G₁, ...
- Three strains of green-seeded lentil plants appear to have the same phenotype. The strains are designated G₁, ...
- Three strains of green-seeded lentil plants appear to have the same phenotype. The strains are designated G₁, ...
- Three strains of green-seeded lentil plants appear to have the same phenotype. The strains are designated G₁, ...
- Three strains of green-seeded lentil plants appear to have the same phenotype. The strains are designated G₁, ...
- Blue flower color is produced in a species of morning glories when dominant alleles are present at two gene lo...
- Blue flower color is produced in a species of morning glories when dominant alleles are present at two gene lo...
- Blue flower color is produced in a species of morning glories when dominant alleles are present at two gene lo...
- The crosses shown are performed between morning glories whose flower color is determined as described in Probl...
- Two pure-breeding strains of summer squash producing yellow fruit, Y₁ and Y₂, are each crossed to a pure-breed...
- Two pure-breeding strains of summer squash producing yellow fruit, Y₁ and Y₂, are each crossed to a pure-breed...
- Two pure-breeding strains of summer squash producing yellow fruit, Y₁ and Y₂, are each crossed to a pure-breed...
- Human ABO blood type is determined by three alleles, two of which (I^A and I^B) produce gene products that mod...
- Epistatic gene interaction results in a modification of the F₂ dihybrid ratio.Give two examples of modified F₂...
- Epistatic gene interaction results in a modification of the F₂ dihybrid ratio.What genetic principle is the ba...
- Epistatic gene interaction results in a modification of the F₂ dihybrid ratio.What is the expected F₂ ratio?