Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
21. Population Genetics
Allelic Frequency Changes
Problem 34a
Textbook Question
Textbook QuestionEvaluate the following pedigree, and answer the questions below. Calculate F for any inbred members of this family.
 Verified Solution
Verified SolutionThis video solution was recommended by our tutors as helpful for the problem above
Video duration:
2mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Inbreeding Coefficient (F)
The inbreeding coefficient (F) quantifies the probability that two alleles at a locus in an individual are identical by descent. It is calculated based on the pedigree of the individual, considering the relatedness of the ancestors. A higher F value indicates a greater degree of inbreeding, which can lead to an increased risk of genetic disorders due to the expression of deleterious recessive alleles.
Recommended video:
Guided course

F Factor and Hfr
Pedigree Analysis
Pedigree analysis is a method used to study the inheritance patterns of traits across generations in a family. It involves constructing a family tree that illustrates the relationships between individuals and their phenotypes. This analysis helps identify carriers of genetic traits and assess the likelihood of traits being passed on, which is essential for calculating inbreeding coefficients.
Recommended video:
Guided course

Pedigree Flowchart
Identical By Descent (IBD)
Identical by descent (IBD) refers to alleles that are inherited from a common ancestor without any intervening mutations. In the context of inbreeding, IBD alleles increase the likelihood of homozygosity for certain traits, which can have significant implications for genetic health. Understanding IBD is crucial for calculating the inbreeding coefficient and assessing the genetic risks associated with inbreeding.
Recommended video:
Guided course

Twin Studies

 5:58m
5:58mWatch next
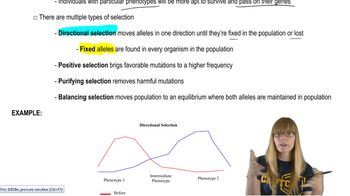
Master Natural Selection with a bite sized video explanation from Kylia Goodner
Start learning