Table of contents
- 1. Introduction to Genetics51m
- 2. Mendel's Laws of Inheritance3h 37m
- 3. Extensions to Mendelian Inheritance2h 41m
- 4. Genetic Mapping and Linkage2h 28m
- 5. Genetics of Bacteria and Viruses1h 21m
- 6. Chromosomal Variation1h 48m
- 7. DNA and Chromosome Structure56m
- 8. DNA Replication1h 10m
- 9. Mitosis and Meiosis1h 34m
- 10. Transcription1h 0m
- 11. Translation58m
- 12. Gene Regulation in Prokaryotes1h 19m
- 13. Gene Regulation in Eukaryotes44m
- 14. Genetic Control of Development44m
- 15. Genomes and Genomics1h 50m
- 16. Transposable Elements47m
- 17. Mutation, Repair, and Recombination1h 6m
- 18. Molecular Genetic Tools19m
- 19. Cancer Genetics29m
- 20. Quantitative Genetics1h 26m
- 21. Population Genetics50m
- 22. Evolutionary Genetics29m
12. Gene Regulation in Prokaryotes
Lac Operon
Problem 21
Textbook Question
Four independent lac⁻ mutants (mutants A to D) are isolated in haploid strains of E. coli. The strains have the following phenotypic characteristics:
Mutant A is lac⁻, but transcription1 of operon genes is induced by lactose.
Mutant B is lac⁻ and has uninducible2 transcription of operon genes.
Mutant C is lac⁺ and has constitutive3 transcription of operon genes.
Mutant D is lac⁺ and has constitutive3 transcription of operon genes.
A microbiologist develops donor and recipient varieties of each mutant strain and crosses them with the results shown below. The table indicates whether inducible, constitutive, or noninducible transcription occurs, along with and growth habit for each partial diploid. Assume each strain has a single mutation.
Mating Transcription and Growth
A × B lac⁻
A × C lac⁺, inducible
A × D lac⁺, constitutive
B × C lac⁺, inducible
B × D lac⁺, constitutive
C × D lac⁺, constitutive
Use this information to identify which lac operon gene is mutated in each strain.
 Verified step by step guidance
Verified step by step guidance1
Step 1: Understand the lac operon system. The lac operon in E. coli is a set of genes involved in lactose metabolism. It includes structural genes (lacZ, lacY, lacA) and regulatory elements (promoter, operator, and the lacI gene encoding the repressor). Mutations in these components can affect transcription in different ways: inducible (normal regulation), constitutive (always on), or noninducible (always off).
Step 2: Analyze the phenotypes of the mutants. Mutant A is lac⁻ but inducible, suggesting a mutation in a structural gene (e.g., lacZ or lacY) that does not affect regulation. Mutant B is lac⁻ and uninducible, indicating a mutation in a regulatory element that prevents transcription entirely (e.g., lacP or lacO). Mutants C and D are lac⁺ and constitutive, suggesting mutations that disrupt repression (e.g., lacI or lacO).
Step 3: Examine the results of the partial diploid matings. For example, in the A × C cross, the phenotype is lac⁺ and inducible. This suggests that the mutation in A is complemented by the wild-type allele in C, and the mutation in C does not affect inducibility. This points to A having a structural gene mutation and C having a regulatory mutation (e.g., lacI or lacO).
Step 4: Use the complementation test results to assign mutations. For instance, in the B × C cross, the phenotype is lac⁺ and inducible. This indicates that the mutation in B is complemented by the wild-type allele in C, and the mutation in C does not prevent inducibility. This suggests B has a mutation in a regulatory element (e.g., lacP) and C has a mutation in lacI or lacO.
Step 5: Summarize the mutations. Based on the data, Mutant A likely has a structural gene mutation (e.g., lacZ or lacY), Mutant B has a mutation in the promoter (lacP), Mutant C has a mutation in the repressor (lacI), and Mutant D has a mutation in the operator (lacO). These assignments are consistent with the phenotypes and complementation test results.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
5mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Lac Operon Structure and Function
The lac operon is a set of genes in E. coli that are involved in the metabolism of lactose. It consists of three structural genes (lacZ, lacY, and lacA) and regulatory elements that control their expression. The operon is typically off but can be induced by lactose, which binds to the repressor protein, allowing transcription of the genes necessary for lactose utilization.
Recommended video:
Guided course
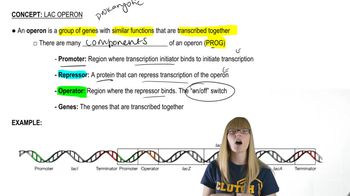
Lac Operon Overview
Mutations and Their Effects
Mutations in the lac operon can lead to different phenotypes, such as lac⁻ (non-functional) or lac⁺ (functional). Specific mutations can affect the repressor's ability to bind to the operator or alter the promoter's activity, resulting in uninducible or constitutive expression of the operon. Understanding these mutations is crucial for determining the functional status of each mutant strain.
Recommended video:
Guided course

Maternal Effect
Partial Diploids and Complementation
In genetic studies, partial diploids are created by introducing a second copy of a gene into a haploid organism, allowing researchers to study gene interactions. In the context of the lac operon, crossing different mutants can reveal whether a mutation is dominant or recessive, and whether the mutations can complement each other to restore function, which is essential for identifying the specific mutations in each strain.
Recommended video:
Guided course

Complementation
Related Videos
Related Practice
Textbook Question
In a theoretical operon, genes A, B, C, and D represent the repressor gene, the promoter sequence, the operator gene, and the structural gene, but not necessarily in the order named. This operon is concerned with the metabolism of a theoretical molecule (tm). From the data provided in the accompanying table, first decide whether the operon is inducible or repressible. Then assign A, B, C, and D to the four parts of the operon. Explain your rationale.(AE=activeenzyme;IE=inactiveenzyme;NE=noenzyme.)Genotype tm Present tm AbsentA⁺B⁺C⁺D⁺ AE NEA⁻B⁺C⁺D⁺ AE AEA⁺B⁻C⁺D⁺ NE NEA⁺B⁺C⁻D⁺ IE NEA⁺B⁺C⁺D⁻ AE AEA⁻B⁺C⁺D⁺/F'A⁺B⁺C⁺D⁺ AE AEA⁺B⁻C⁺D⁺/F'A⁺B⁺C⁺D⁺ AE NEA⁺B⁺C⁻D⁺/F'A⁺B⁺C⁺D⁺ AE+IE NEA⁺B⁺C⁺D⁻/F'A⁺B⁺C⁺D⁺ AE NE
1140
views


