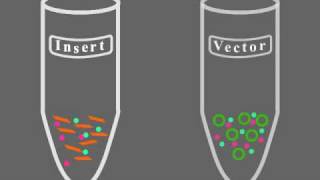
Hey there, this is Eric Simon from New England College in Henniker, New Hampshire. In this Visualizing the Concept video we'll walk through the process of creating recombinant DNA. Let's get started. Many genetic engineering experiments involve combining DNA from more than one source often from different species. For example, a human gene encoding for an important protein may be inserted into a bacterium. Such a modified bacterium will then reproduce and express the human protein which can then be harvested in large quantities perhaps by a pharmaceutical company to produce a new drug. In another example, an insect gene might be placed into a plant in order to improve that plant's ability to resist cold weather. To create recombinant DNA scientists must cut pieces of DNA from multiple sources and then past them together into a single recombinant DNA molecule. In this animation we're going to focus on the actual cutting and pasting of DNA to see how a typical genetic engineering project is undertaken. To help visualize this process let's use household materials. Imagine that you have a necklace made from blue ribbon and you wish to insert a bit of red ribbon into it. To do this you would snip out a piece of the red ribbon by making two cuts, then open up the blue necklace by making one cut. You could then insert the new ribbon and seal the gaps. You now have a recombinant necklace that contains pieces from two different ribbons combined together. To accomplish a similar process with DNA genetic engineers often start with plasmids, small circular pieces of DNA found within many bacteria such as this E. coli. Once a researcher has removed a plasmid from a bacterium it can be manipulated in the laboratory. In this experiment the circular plasmid needs to be cut open. This is accomplished using a restriction enzyme, a protein that cuts DNA at one specific sequence of bases. The restriction enzyme shown here for example is called EcoR1. It cuts DNA at one specific sequence of bases called the restriction site. For example, the restriction enzyme EcoR will only cut DNA at the sequence G-A-A-T-T-C. When this restriction enzyme is added to the plasma DNA it will cut the DNA at every location that contains the sequence and no places that don't. The specific restriction enzyme used in the genetic engineering project varies from experiment to experiment, but an engineer will usually choose a restriction enzyme that will cut the bacterial plasmid in just one location opening up the circle. If a plasmid contains two restriction sites for a particular restriction enzyme how many separate pieces of DNA will result? The result of cutting a circular plasmid twice will be two linear pieces of DNA. Imagine snipping a rubber band twice, you'll be left with two pieces. Once the bacterial plasmid is snipped open it's ready to have another piece of DNA inserted into it. Here you can see a human cell that contains a target gene. Once the DNA is extracted from the cell the same restriction enzyme can be used to cut it up. Because the human chromosome is so big it will probably be cut into several pieces, one of which contains our target gene shown in red. If a linear piece of DNA contains two restriction sites for a particular restriction enzyme how many separate pieces of DNA will result? In this case since the starting DNA is linear two cuts will result in three linear pieces of DNA. Imagine cutting a strip of paper twice, you'd be left holding three smaller strips of paper. Now that we have our two desired segments of DNA, one bacterial plasmid and one human gene, let's see how they're pasted together. The key is that the restriction enzyme used did not cut across the DNA double helix evenly. Instead it left staggered cuts called sticky ends. Each sticky end is a single stranded set of unpaired DNA bases. The bases in the single stranded regions are no longer hydrogen bonded to their matching bases on the other strand. These unpaired bases are now available to pair up with a complementary set of bases on another sticky end. Notice that the DNA base sequences of the sticky ends on the bacterial plasmid DNA and the human DNA match up. The key to creating recombinant DNA is to use the same restriction enzyme to cut both the plasmid DNA and the target DNA. Because the same enzyme was used the same sequences were cut in the same way. As a consequence the unpaired based sequences are complementary to one another and the sticky end of one piece of DNA will match up with the sticky end from another. The hydrogen bonds that form between the sticky ends of the two pieces of DNA temporarily hold the foreign DNA in the middle of the plasmid. To permanently pasted these separate pieces of DNA the engineer can add a DNA pasting enzyme called DNA ligase. This enzyme builds new covalent bonds sealing up the separate pieces of DNA into a single recombinant piece of DNA. The goal of creating recombinant DNA has now been achieved. Once created, the recombinant plasmid can be inserted into a bacterium. The recombinant plasmid will multiply to produce a colony of identical bacteria, each of which carries the foreign gene in the recombinant plasmid. As it goes about its life cycle a recombinant bacterium will treat the foreign DNA as if it were its own. This means that the foreign DNA will be transcribed into an RNA molecule and then translated into a protein. Perhaps the protein is a pharmaceutical product that can serve as a drug. Right now in the U.S. Mid-West there are large tanks of recombinant bacteria carrying the gene for human insulin. The bacteria transcribe and translate the gene to produce human insulin protein which can then be extracted and purified. This human insulin is then packaged and sold to millions of diabetics for whom this drug made from recombinant DNA is a life-saving medication.
Table of contents
- 1. Introduction to Biology2h 40m
- 2. Chemistry3h 40m
- 3. Water1h 26m
- 4. Biomolecules2h 23m
- 5. Cell Components2h 26m
- 6. The Membrane2h 31m
- 7. Energy and Metabolism2h 0m
- 8. Respiration2h 40m
- 9. Photosynthesis2h 49m
- 10. Cell Signaling59m
- 11. Cell Division2h 47m
- 12. Meiosis2h 0m
- 13. Mendelian Genetics4h 41m
- Introduction to Mendel's Experiments7m
- Genotype vs. Phenotype17m
- Punnett Squares13m
- Mendel's Experiments26m
- Mendel's Laws18m
- Monohybrid Crosses16m
- Test Crosses14m
- Dihybrid Crosses20m
- Punnett Square Probability26m
- Incomplete Dominance vs. Codominance20m
- Epistasis7m
- Non-Mendelian Genetics12m
- Pedigrees6m
- Autosomal Inheritance21m
- Sex-Linked Inheritance43m
- X-Inactivation9m
- 14. DNA Synthesis2h 27m
- 15. Gene Expression3h 20m
- 16. Regulation of Expression3h 31m
- Introduction to Regulation of Gene Expression13m
- Prokaryotic Gene Regulation via Operons27m
- The Lac Operon21m
- Glucose's Impact on Lac Operon25m
- The Trp Operon20m
- Review of the Lac Operon & Trp Operon11m
- Introduction to Eukaryotic Gene Regulation9m
- Eukaryotic Chromatin Modifications16m
- Eukaryotic Transcriptional Control22m
- Eukaryotic Post-Transcriptional Regulation28m
- Eukaryotic Post-Translational Regulation13m
- 17. Viruses37m
- 18. Biotechnology2h 58m
- 19. Genomics17m
- 20. Development1h 5m
- 21. Evolution3h 1m
- 22. Evolution of Populations3h 52m
- 23. Speciation1h 37m
- 24. History of Life on Earth2h 6m
- 25. Phylogeny40m
- 26. Prokaryotes4h 59m
- 27. Protists1h 6m
- 28. Plants1h 22m
- 29. Fungi36m
- 30. Overview of Animals34m
- 31. Invertebrates1h 2m
- 32. Vertebrates50m
- 33. Plant Anatomy1h 3m
- 34. Vascular Plant Transport2m
- 35. Soil37m
- 36. Plant Reproduction47m
- 37. Plant Sensation and Response1h 9m
- 38. Animal Form and Function1h 19m
- 39. Digestive System10m
- 40. Circulatory System1h 57m
- 41. Immune System1h 12m
- 42. Osmoregulation and Excretion50m
- 43. Endocrine System4m
- 44. Animal Reproduction2m
- 45. Nervous System55m
- 46. Sensory Systems46m
- 47. Muscle Systems23m
- 48. Ecology3h 11m
- Introduction to Ecology20m
- Biogeography14m
- Earth's Climate Patterns50m
- Introduction to Terrestrial Biomes10m
- Terrestrial Biomes: Near Equator13m
- Terrestrial Biomes: Temperate Regions10m
- Terrestrial Biomes: Northern Regions15m
- Introduction to Aquatic Biomes27m
- Freshwater Aquatic Biomes14m
- Marine Aquatic Biomes13m
- 49. Animal Behavior28m
- 50. Population Ecology3h 41m
- Introduction to Population Ecology28m
- Population Sampling Methods23m
- Life History12m
- Population Demography17m
- Factors Limiting Population Growth14m
- Introduction to Population Growth Models22m
- Linear Population Growth6m
- Exponential Population Growth29m
- Logistic Population Growth32m
- r/K Selection10m
- The Human Population22m
- 51. Community Ecology2h 46m
- Introduction to Community Ecology2m
- Introduction to Community Interactions9m
- Community Interactions: Competition (-/-)38m
- Community Interactions: Exploitation (+/-)23m
- Community Interactions: Mutualism (+/+) & Commensalism (+/0)9m
- Community Structure35m
- Community Dynamics26m
- Geographic Impact on Communities21m
- 52. Ecosystems2h 36m
- 53. Conservation Biology24m
18. Biotechnology
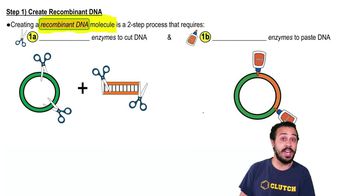
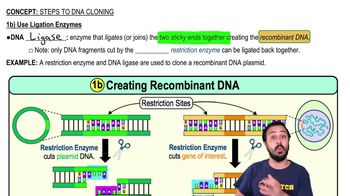
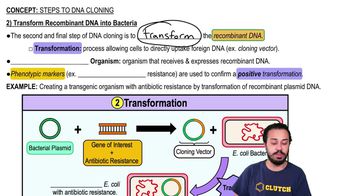
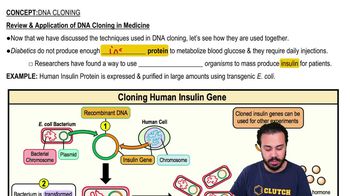
Steps to DNA Cloning
Video duration:
7mPlay a video:
Related Videos
Related Practice





![[LECT C8 : RECOMBINANT DNA TECHNOLOGY] 8.2 Steps in Gene Cloning](https://img.youtube.com/vi/T1l-StLrDwg/mqdefault.jpg)