Table of contents
- 1. Introduction to Biology2h 42m
- 2. Chemistry3h 40m
- 3. Water1h 26m
- 4. Biomolecules2h 23m
- 5. Cell Components2h 26m
- 6. The Membrane2h 31m
- 7. Energy and Metabolism2h 0m
- 8. Respiration2h 40m
- 9. Photosynthesis2h 49m
- 10. Cell Signaling59m
- 11. Cell Division2h 47m
- 12. Meiosis2h 0m
- 13. Mendelian Genetics4h 44m
- Introduction to Mendel's Experiments7m
- Genotype vs. Phenotype17m
- Punnett Squares13m
- Mendel's Experiments26m
- Mendel's Laws18m
- Monohybrid Crosses19m
- Test Crosses14m
- Dihybrid Crosses20m
- Punnett Square Probability26m
- Incomplete Dominance vs. Codominance20m
- Epistasis7m
- Non-Mendelian Genetics12m
- Pedigrees6m
- Autosomal Inheritance21m
- Sex-Linked Inheritance43m
- X-Inactivation9m
- 14. DNA Synthesis2h 27m
- 15. Gene Expression3h 20m
- 16. Regulation of Expression3h 31m
- Introduction to Regulation of Gene Expression13m
- Prokaryotic Gene Regulation via Operons27m
- The Lac Operon21m
- Glucose's Impact on Lac Operon25m
- The Trp Operon20m
- Review of the Lac Operon & Trp Operon11m
- Introduction to Eukaryotic Gene Regulation9m
- Eukaryotic Chromatin Modifications16m
- Eukaryotic Transcriptional Control22m
- Eukaryotic Post-Transcriptional Regulation28m
- Eukaryotic Post-Translational Regulation13m
- 17. Viruses37m
- 18. Biotechnology2h 58m
- 19. Genomics17m
- 20. Development1h 5m
- 21. Evolution3h 1m
- 22. Evolution of Populations3h 52m
- 23. Speciation1h 37m
- 24. History of Life on Earth2h 6m
- 25. Phylogeny2h 31m
- 26. Prokaryotes4h 59m
- 27. Protists1h 12m
- 28. Plants1h 22m
- 29. Fungi36m
- 30. Overview of Animals34m
- 31. Invertebrates1h 2m
- 32. Vertebrates50m
- 33. Plant Anatomy1h 3m
- 34. Vascular Plant Transport1h 2m
- 35. Soil37m
- 36. Plant Reproduction47m
- 37. Plant Sensation and Response1h 9m
- 38. Animal Form and Function1h 19m
- 39. Digestive System1h 10m
- 40. Circulatory System1h 57m
- 41. Immune System1h 12m
- 42. Osmoregulation and Excretion50m
- 43. Endocrine System1h 4m
- 44. Animal Reproduction1h 2m
- 45. Nervous System1h 55m
- 46. Sensory Systems46m
- 47. Muscle Systems23m
- 48. Ecology3h 11m
- Introduction to Ecology20m
- Biogeography14m
- Earth's Climate Patterns50m
- Introduction to Terrestrial Biomes10m
- Terrestrial Biomes: Near Equator13m
- Terrestrial Biomes: Temperate Regions10m
- Terrestrial Biomes: Northern Regions15m
- Introduction to Aquatic Biomes27m
- Freshwater Aquatic Biomes14m
- Marine Aquatic Biomes13m
- 49. Animal Behavior28m
- 50. Population Ecology3h 41m
- Introduction to Population Ecology28m
- Population Sampling Methods23m
- Life History12m
- Population Demography17m
- Factors Limiting Population Growth14m
- Introduction to Population Growth Models22m
- Linear Population Growth6m
- Exponential Population Growth29m
- Logistic Population Growth32m
- r/K Selection10m
- The Human Population22m
- 51. Community Ecology2h 46m
- Introduction to Community Ecology2m
- Introduction to Community Interactions9m
- Community Interactions: Competition (-/-)38m
- Community Interactions: Exploitation (+/-)23m
- Community Interactions: Mutualism (+/+) & Commensalism (+/0)9m
- Community Structure35m
- Community Dynamics26m
- Geographic Impact on Communities21m
- 52. Ecosystems2h 36m
- 53. Conservation Biology24m
18. Biotechnology
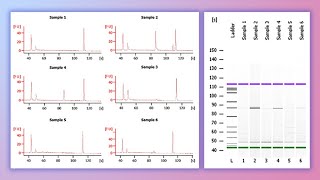
DNA Fingerprinting
Problem 4`
Textbook Question
A paleontologist has recovered a bit of tissue from the 400-year-old preserved skin of an extinct dodo (a bird). To compare a specific region of the DNA from a sample with DNA from living birds, which of the following would be most useful for increasing the amount of dodo DNA available for testing?
a. SNP analysis
b. Polymerase chain reaction (PCR)
c. Electroporation
d. Gel electrophoresis
 Verified step by step guidance
Verified step by step guidance1
Understand the problem: The goal is to increase the amount of dodo DNA available for testing. This requires a method that can amplify small amounts of DNA.
Review the options: a. SNP analysis is used for identifying variations in DNA sequences, not for amplification. b. Polymerase chain reaction (PCR) is a technique specifically designed to amplify DNA. c. Electroporation is a method to introduce DNA into cells, not for amplification. d. Gel electrophoresis is used for separating DNA fragments by size, not for amplification.
Focus on PCR: Polymerase chain reaction (PCR) is a technique that can exponentially amplify a specific DNA segment. It involves repeated cycles of denaturation, annealing, and extension.
Explain PCR process: In PCR, the DNA sample is first denatured by heating to separate the strands. Then, primers anneal to the target DNA sequence. DNA polymerase extends the primers, synthesizing new DNA strands. This cycle is repeated multiple times to amplify the DNA.
Conclude: Based on the need to increase the amount of DNA, PCR is the most suitable method for amplifying the dodo DNA for further testing and comparison with living birds.
 Verified video answer for a similar problem:
Verified video answer for a similar problem:This video solution was recommended by our tutors as helpful for the problem above
Video duration:
1mPlay a video:
Was this helpful?
Key Concepts
Here are the essential concepts you must grasp in order to answer the question correctly.
Polymerase Chain Reaction (PCR)
Polymerase Chain Reaction (PCR) is a technique used to amplify small segments of DNA, making it possible to generate thousands to millions of copies of a particular DNA sequence. This is crucial when working with limited or degraded DNA samples, such as those from extinct species, as it allows for sufficient material to be available for further analysis and comparison.
Recommended video:
Guided course

Introduction to Polymerase Chain Reaction
DNA Amplification
DNA amplification is the process of creating multiple copies of a specific DNA sequence. This is essential in genetic studies, especially when the initial sample is too small for analysis. Techniques like PCR are employed to amplify DNA, enabling researchers to conduct detailed studies on genetic material that would otherwise be too scarce to examine.
Recommended video:
Guided course

Signal Amplification
Comparative Genomics
Comparative genomics involves comparing the DNA sequences of different species to understand their evolutionary relationships and functional biology. By analyzing the DNA of extinct species like the dodo and comparing it with living birds, scientists can gain insights into evolutionary changes, genetic diversity, and the functional aspects of genes across species.
Recommended video:
Guided course

Genomes and Genome Evolution
Related Videos
Related Practice