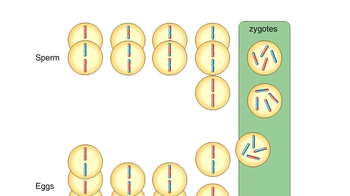
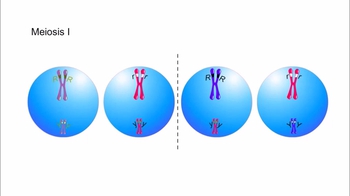
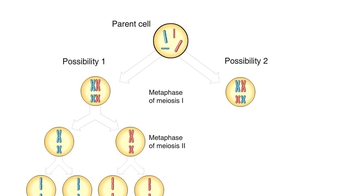
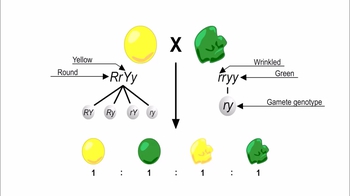
Let’s examine how genetic crosses are analyzed to determine whether genes are linked and to determine the map distance between linked genes on a chromosome. In this cross we will look at the characters of eye shape and tooth number in MendAliens, a mythical alien species. We begin with true-breeding parental aliens. One parent is homozygous for round eyes and no tooth. The other parent is homozygous for vertical eyes and one tooth. Let’s make the cross. Note that the F1 dihybrid offspring have round eyes and no tooth. The simplest hypothesis for now is that there are two genes for these two characters, with the round-eyes allele dominant to the vertical-eyes allele and the no-tooth allele dominant to the one-tooth allele. Now, let’s see what happens when an F1 dihybrid is testcrossed with a homozygous recessive individual, an alien with vertical eyes and one tooth. Here are the results of the testcross. If the two genes showed independent assortment, we would expect a ratio of 1 to 1 to 1 to in the phenotypes from the testcross. Instead, most offspring resemble the phenotypes of the parents from the original P generation; that is, the parent with round eyes and no tooth, and the parent with vertical eyes and one tooth. We can conclude that the genes for eye shape and tooth number do not assort independently. Instead, the eye-shape and tooth-number alleles are usually inherited together because the genes for these two characters are near each other on the same chromosome. Such genes are said to be linked. For linked genes, we need to include chromosomes in our genotype diagrams, as shown here. We’ll use capital R for the round-eyes allele and small r for the vertical-eyes allele, and capital N for the no-tooth allele and small n for the one-tooth allele. The parent with round eyes and no tooth has two homologous chromosomes with capital R and capital N on each chromosome. This parent can produce only one type of gamete, which has a chromosome with capital R and capital N on the same chromosome. Similarly, the parent with vertical eyes and one tooth can produce only gametes with small r and small n on the same chromosome. The F1 dihybrids inherit one chromosome with capital R and capital N from the parent with round eyes and no tooth, and one chromosome with small r and small n from the parent with vertical eyes and one tooth. These two chromosomes are homologous. Since the round-eyes allele (capital R) is dominant, the F1 dihybrids have round eyes. And since the no-tooth allele (capital N) is dominant, the F1 dihybrids have no tooth. The F1 dihybrids can produce four types of gametes during meiosis. If crossing over does not occur between the homologous chromosomes during meiosis, gametes are produced that have either the dominant alleles for both characters (capital R and capital N) or the recessive alleles for both characters (small r and small n). These gametes have the same alleles as the gametes from the original P generation. If crossing over does occur between the homologous chromosomes during gamete formation, recombinant chromosomes are produced with either capital R and small n or small r and capital N. These gametes have combinations of alleles not seen before in either the P generation or the F1 dihybrids. Now let's reexamine the testcross between an F1 dihybrid and a homozygous recessive individual, an alien with vertical eyes and one tooth. The homozygous recessive MendAlien can produce only one type of gamete, which has small r and small n on the same chromosome because even if crossing over occurred, it would just be swapping the same alleles. This Punnett square shows the genotype and phenotype of each type of offspring. Notice that there are many more parental-type offspring than recombinant offspring, indicating that these two genes did not assort independently, but instead are near each other on the same chromosome and are therefore linked. Let's look at a summary of the offspring from the testcross. We can calculate the frequency of recombinant offspring by using the following formula: recombination frequency equals number of recombinants divided by the total offspring times 100. In this example, the recombination frequency equals 25 recombinants divided by 175 total offspring times 100, which equals 14.3 percent. The recombination frequency is directly related to how far apart two genes are on a chromosome. Generally speaking, genes that are closer together, such as the eye-shape gene and the tooth-number gene, are less likely to have a crossover occur between them. Genes that are farther apart, such as the eye-shape gene and the ear-shape gene, have a greater chance of a crossover occurring between them. This tendency is reflected in a higher frequency of recombinant offspring in genes that are farther apart. Therefore, recombination frequency data are used to assign gene positions on a chromosome map. By definition, one map unit is equivalent to a 1 percent recombination frequency. Thus, the genes for eye shape and tooth number are 14.3 map units apart. In a separate experiment, it was determined that the recombination frequency for the eye-shape gene and the ear-shape gene is 21 percent. Thus, the genes for eye shape and ear shape are 21 map units apart.
Table of contents
- 1. Introduction to Biology2h 40m
- 2. Chemistry3h 40m
- 3. Water1h 26m
- 4. Biomolecules2h 23m
- 5. Cell Components2h 26m
- 6. The Membrane2h 31m
- 7. Energy and Metabolism2h 0m
- 8. Respiration2h 40m
- 9. Photosynthesis2h 49m
- 10. Cell Signaling59m
- 11. Cell Division2h 47m
- 12. Meiosis2h 0m
- 13. Mendelian Genetics4h 41m
- Introduction to Mendel's Experiments7m
- Genotype vs. Phenotype17m
- Punnett Squares13m
- Mendel's Experiments26m
- Mendel's Laws18m
- Monohybrid Crosses16m
- Test Crosses14m
- Dihybrid Crosses20m
- Punnett Square Probability26m
- Incomplete Dominance vs. Codominance20m
- Epistasis7m
- Non-Mendelian Genetics12m
- Pedigrees6m
- Autosomal Inheritance21m
- Sex-Linked Inheritance43m
- X-Inactivation9m
- 14. DNA Synthesis2h 27m
- 15. Gene Expression3h 20m
- 16. Regulation of Expression3h 31m
- Introduction to Regulation of Gene Expression13m
- Prokaryotic Gene Regulation via Operons27m
- The Lac Operon21m
- Glucose's Impact on Lac Operon25m
- The Trp Operon20m
- Review of the Lac Operon & Trp Operon11m
- Introduction to Eukaryotic Gene Regulation9m
- Eukaryotic Chromatin Modifications16m
- Eukaryotic Transcriptional Control22m
- Eukaryotic Post-Transcriptional Regulation28m
- Eukaryotic Post-Translational Regulation13m
- 17. Viruses37m
- 18. Biotechnology2h 58m
- 19. Genomics17m
- 20. Development1h 5m
- 21. Evolution3h 1m
- 22. Evolution of Populations3h 52m
- 23. Speciation1h 37m
- 24. History of Life on Earth2h 6m
- 25. Phylogeny2h 31m
- 26. Prokaryotes4h 59m
- 27. Protists1h 12m
- 28. Plants1h 22m
- 29. Fungi36m
- 30. Overview of Animals34m
- 31. Invertebrates1h 2m
- 32. Vertebrates50m
- 33. Plant Anatomy1h 3m
- 34. Vascular Plant Transport2m
- 35. Soil37m
- 36. Plant Reproduction47m
- 37. Plant Sensation and Response1h 9m
- 38. Animal Form and Function1h 19m
- 39. Digestive System10m
- 40. Circulatory System1h 57m
- 41. Immune System1h 12m
- 42. Osmoregulation and Excretion50m
- 43. Endocrine System4m
- 44. Animal Reproduction2m
- 45. Nervous System55m
- 46. Sensory Systems46m
- 47. Muscle Systems23m
- 48. Ecology3h 11m
- Introduction to Ecology20m
- Biogeography14m
- Earth's Climate Patterns50m
- Introduction to Terrestrial Biomes10m
- Terrestrial Biomes: Near Equator13m
- Terrestrial Biomes: Temperate Regions10m
- Terrestrial Biomes: Northern Regions15m
- Introduction to Aquatic Biomes27m
- Freshwater Aquatic Biomes14m
- Marine Aquatic Biomes13m
- 49. Animal Behavior28m
- 50. Population Ecology3h 41m
- Introduction to Population Ecology28m
- Population Sampling Methods23m
- Life History12m
- Population Demography17m
- Factors Limiting Population Growth14m
- Introduction to Population Growth Models22m
- Linear Population Growth6m
- Exponential Population Growth29m
- Logistic Population Growth32m
- r/K Selection10m
- The Human Population22m
- 51. Community Ecology2h 46m
- Introduction to Community Ecology2m
- Introduction to Community Interactions9m
- Community Interactions: Competition (-/-)38m
- Community Interactions: Exploitation (+/-)23m
- Community Interactions: Mutualism (+/+) & Commensalism (+/0)9m
- Community Structure35m
- Community Dynamics26m
- Geographic Impact on Communities21m
- 52. Ecosystems2h 36m
- 53. Conservation Biology24m
12. Meiosis
Genetic Variation During Meiosis
Video duration:
6mPlay a video:
Related Videos
Related Practice