In this video, we're going to talk about how to calculate the isoelectric point of a peptide. So, recall from our previous lesson videos on the isoelectric point that the isoelectric point, or the PI calculation, is always just going to be the midpoint or the average between the 2 pKa values for the 2 ionizations involving the neutral species. And so, all this is saying is that, whenever we're calculating the isoelectric point, we're only going to average 2 pKa values, regardless of how many pKa values were given, and the same is going to apply for calculating the isoelectric point of a peptide. And so, before we get to our example below, there's really 2 ideas that I really want to emphasize. And the first is a similar idea to the one that we talked about when we were calculating the net charge of a peptide. And this is the idea that the isoelectric point and the net charge of a peptide can both only be estimated. They can only be estimated, and this is because of the unique microenvironment that each residue has in a peptide. And so, the true PI values can only be experimentally determined. And because we know that the microenvironment is capable of shifting the pKa values of residues, you need to make sure that you're using the correct set of pKa values, specifically the set of pKa values for amino acid residues and not for pKa values of free amino acids. And so if your professor uses the same set of pKa values for amino acid residues and free amino acids, then you're good to go and you don't have to worry about this. But just in case, you should double-check to make sure that your professor doesn't want you to use a specific set of pKa values for amino acid residues. And so when we're doing our practice problems, we're going to be using a specific set of pKa values for amino acid residues. And so the second idea that I really want to emphasize is just this idea that the isoelectric point of a peptide follows similar steps for calculating the isoelectric point of amino acids with ionizable R groups. And so this little, chart here, this little diagram might look familiar to you because it's really the same one from our previous lesson. And so let's take a look at the 4 steps that we need to follow to calculate the isoelectric point of a peptide. And again, the first step is just going to be to know how the amino acid R groups ionize. And really all this is saying is that we need to know the 7 amino acids with ionizable R groups. And recall that our mnemonic to help us remember those 7 amino acids is just "yucky crazy dragons knights riding horses". And so if you know this mnemonic and how it works, then you'll know those 7 amino acid, with ionizable R groups and you'll be set to go with step number 1. Now step number 2 is going to be to order the pKa values from the smallest to the greatest. And so here, we're showing only 4 pKa values, but regardless of how many pKa values there are, there could be 5, there could be 6, there could be 8, however many there are, you're gonna want to, order them all from the smallest to the largest. And so step number 3 is going to be to determine the net charge of predominant structures at any pH between each pair of adjacent pKa values or neighboring pKa values. And so what you can see down here is that we've got our 4 pKa values ordered from smallest to greatest and then, we have these, pH ranges in between the pKa values that we're going to check for the predominant structure. And we can either imagine the predominant structure, so we don't need to draw it out unless it tells us to. Or if you're still working on, if you're having trouble imagining it, you can always draw the predominant structures. But the whole point is just to determine the net charges of the predominant structures, and so we've got these ranges in between each pKa that we need to check. And so, we would check each one of these. And so the way that you determine the net charge, again, of the predominant structure, is by comparing the pH of the solution that we choose to the pKa values of all the ionizable groups. And of course, the last step is going to be to calculate the isoelectric point just by averaging the 2 pKa values, so it's always just going to be 2. It's gonna be the 2 pKa values that sandwich the pH where the predominant structure has a neutral net charge. So again, the isoelectric point is going to be the pH where the structure is going to have a neutral net charge or a net charge of 0. And so whatever 2 pKa values they are, so if it were this one and this one, you would average these 2 pKa values if this structure here had a neutral net charge. If it wasn't these two pKa values, if it was this pK and this pKa because this structure had a neutral net charge, then you would average these 2 pKa values here. So it's all about where do you find that neutral net charge. And so, let's go ahead and move on to our example. And in this example, it says to estimate the isoelectric point for the following peptide, DGE, which is aspartic acid, glycine, and glutamic acid. And so, all we're gonna do here is consider all of the ionizable groups. So we don't really care about any of the other groups that are not ionizable because they're not gonna contribute to the net charge, and so if they don't contribute to the net charge, then it's not gonna contribute to the isoelectric point. And so for D, G, and E, we can rewrite it over here. And so we basically got a few ionizable groups that we need to consider. We know that the first amino acid residue in a chain is gonna have an ionizable amino group, so there's an ionizable amino group that we need to consider. Now, aspartic acid is going to be one of our negatively charged acetic amino acid R groups. So we know that its r group potentially could have a negative charge, so we'll also leave it as a blank here to consider its charge. Now, glycine does not have an ionizable R group. It's not one of our 7 amino acids with ionizable R-groups, so we don't care about its R group and it doesn't have any ionizable amino or carboxyl groups because it's an internal amino acid residue. Now, glutamic acid over here does have an ionizable R group that's potentially negative, so we are going to consider that. And then also because it's the last residue in the chain, it also has a potentially negative carboxyl group. And so all we need to do is, now that we've done step number 1, we know how these amino acid R groups ionize and we've even highlighted them like this to consider their ionizations. Step number 2 is gonna be to order the pKa values for all 4 of these ionizable groups in order from smallest to largest. So now, we're going to use our pKa set over here for amino acid residues, and we'll start with the amino group over here, and the amino group is gonna be the N-terminus. And so, notice that the N-terminus has a pKa of 8, and the pKa of 8 is, when we compare it to all of the other pKa values that we're going to be looking at, we have glutamic acid, for glutamic acid, has a pKa of 4.1. We have aspartic acid, which is the D here, has a pKa of 3.9. And then we also have the carboxyl group, and the carboxyl group is the C-terminus up here, which has a pKa of 3.5. So we have our 4 pKa values, we need to order them from smallest to largest, and so the smallest one is the pKa of the C-terminus. So we'll put 3.5 over here, and we'll put carboxyl above it to remind us it's the carboxyl group. The next one is aspartic acid, so aspartic acid is 3.9 and this is we'll put ASP for aspartic acid to R group. The next one is going to be glutamic acid, which is 4.1. So we'll put 4.1 in a Glu for glutamic acid. And then, of course, our last one's going to be the N-terminus, which is 8.0, and so this is for the amino group. And so, again, all we need to do is choose any range. We can choose this range here or this range here or this range here, it doesn't matter. You can start with anyone that you want. Let's start with the one on the left since we normally do things from left to right anyway. So let's choose this range right here. We can pick any pH that we want as long as it falls into this range, as long as it falls between these 2 pKa values. So let's just choose, I don't know, 3.6, pH of 3.6. And so all we need to do is imagine the predominant structure at pH 3.6 and determine the net charge of the predominant structure at pH 3.6. And so all we need to do is compare the pH to each of the pKa values. So let's start with the amino group. So with the amino group, the pKa is 8. And so a pH of 3.6 is less than the pKa of 8. And when the pKa is greater than the pH, that means that the conjugate acid's going to predominate. And the conjugate acid of the amino group is an NH3+ with a positive charge. So up here, what we're gonna do for our amino group is we're gonna put a plus for the positive charge to consider the charge. So now, let's move on to aspartic acid since that's next in our chain here. Aspartic acid's R group. And so we compare the pH to aspartic acid's R group which is 3.9. And a pH of 3.6 is less than the pH of 3.9, so that means that the conjugate acid form of aspartic acid's R group is going to predominate. And aspartic acid's conjugate acid actually has a neutral charge. So it has a charge of 0. So in here, what we can do is just put a 0 because it has a neutral net charge at pH 3.6. So then we can check, glutamic acid's R group, which is here since it's next in our chain, and glutamic acid's R group is here, its pKa is 4.1. And so, a pH of 3.6 is less than the pH of 4.1. When the pH is less than the pKa, the conjugate acid predominates. The conjugate acid of glutamic acid has a neutral net charge of 0. So we'll just put a 0 in here. And then, we take a look at the carboxyl group, which is last and the carboxyl group, as you can see, which is, way over here, it's pKa is 3.5. And the pKa of 3.5 is less than the pH of 3.6. And when the pH is greater than the pKa, the conjugate base predominates and the conjugate base of the carboxyl group is going to be a carboxylate anion, oops, with a negative charge. We'll put this in blue. And so, all we need to do is total the net charges of each of these ionizable groups to get the net charge of, the structure at pH 3.6. So we have a positive and a negative. So this group over here cancels out with this group over here, and then the other two groups are neutral. So that means that at this pH range, at any pH in here, our structure has a neutral net charge of 0. And so recall that, really when we're calculating the PI, we want to find the, pH range that has a neutral net charge of 0. So we were able to find it on our first try. Now, if we were to choose a different range, if we had selected a different range GCBOVER HERE REMEMBER THAT, WHEN YOU CROSS A pKa, that the net charge of the predominant structure changes by just one unit. And when you go from left to right, it goes down one charge. And so from going from this yellow range over to this green range, and crossing this pKa here, we know that the predominant structure of the net charge is going to change. So any pH in this range, it's going to go down by 1, so we know it would be negative one over here. And so if we had chosen this pH range, then all we would have done is done the same cheat sheet, but we would have gone the opposite direction over here because we know that the charge would go up by 1, and that would give us a net charge of 0. So it doesn't really matter which pH range that you choose. You choose 1, determine the net charge, and then you can use our cheat sheet here to determine the net charge in a different range. So now that we found our neutral net charge, we have to average the 2 pKa values sandwiching the neutral net charge. So the 2 pKa values sandwiching it are this one here and this one here. So we can do our PI calculation over here, and it's just going to be the average of these two numbers. So 3.9 plus 3.5 divided by 2. And so this comes out to 7.4 divided by 2, which comes out to 3.7. And what you'll see is that the pI of 3.7 matches our answer option A. So that means that A here is correct, we can give it a check, indicate that A here is correct, and all our other ones are incorrect, so we can cross them off. So this concludes our example here in our lesson and we'll get some practice calculating the isoelectric point of peptides in our practice. So I'll see you guys in those videos.
- 1. Introduction to Biochemistry4h 34m
- What is Biochemistry?5m
- Characteristics of Life12m
- Abiogenesis13m
- Nucleic Acids16m
- Proteins12m
- Carbohydrates8m
- Lipids10m
- Taxonomy10m
- Cell Organelles12m
- Endosymbiotic Theory11m
- Central Dogma22m
- Functional Groups15m
- Chemical Bonds13m
- Organic Chemistry31m
- Entropy17m
- Second Law of Thermodynamics11m
- Equilibrium Constant10m
- Gibbs Free Energy37m
- 2. Water3h 23m
- 3. Amino Acids8h 10m
- Amino Acid Groups8m
- Amino Acid Three Letter Code13m
- Amino Acid One Letter Code37m
- Amino Acid Configuration20m
- Essential Amino Acids14m
- Nonpolar Amino Acids21m
- Aromatic Amino Acids14m
- Polar Amino Acids16m
- Charged Amino Acids40m
- How to Memorize Amino Acids1h 7m
- Zwitterion33m
- Non-Ionizable Vs. Ionizable R-Groups11m
- Isoelectric Point10m
- Isoelectric Point of Amino Acids with Ionizable R-Groups51m
- Titrations of Amino Acids with Non-Ionizable R-Groups44m
- Titrations of Amino Acids with Ionizable R-Groups38m
- Amino Acids and Henderson-Hasselbalch44m
- 4. Protein Structure10h 4m
- Peptide Bond18m
- Primary Structure of Protein31m
- Altering Primary Protein Structure15m
- Drawing a Peptide44m
- Determining Net Charge of a Peptide42m
- Isoelectric Point of a Peptide37m
- Approximating Protein Mass7m
- Peptide Group22m
- Ramachandran Plot26m
- Atypical Ramachandran Plots12m
- Alpha Helix15m
- Alpha Helix Pitch and Rise20m
- Alpha Helix Hydrogen Bonding24m
- Alpha Helix Disruption23m
- Beta Strand12m
- Beta Sheet12m
- Antiparallel and Parallel Beta Sheets39m
- Beta Turns26m
- Tertiary Structure of Protein16m
- Protein Motifs and Domains23m
- Denaturation14m
- Anfinsen Experiment20m
- Protein Folding34m
- Chaperone Proteins19m
- Prions4m
- Quaternary Structure15m
- Simple Vs. Conjugated Proteins10m
- Fibrous and Globular Proteins11m
- 5. Protein Techniques14h 5m
- Protein Purification7m
- Protein Extraction5m
- Differential Centrifugation15m
- Salting Out18m
- Dialysis9m
- Column Chromatography11m
- Ion-Exchange Chromatography35m
- Anion-Exchange Chromatography38m
- Size Exclusion Chromatography28m
- Affinity Chromatography16m
- Specific Activity16m
- HPLC29m
- Spectrophotometry51m
- Native Gel Electrophoresis23m
- SDS-PAGE34m
- SDS-PAGE Strategies16m
- Isoelectric Focusing17m
- 2D-Electrophoresis23m
- Diagonal Electrophoresis29m
- Mass Spectrometry12m
- Mass Spectrum47m
- Tandem Mass Spectrometry16m
- Peptide Mass Fingerprinting16m
- Overview of Direct Protein Sequencing30m
- Amino Acid Hydrolysis10m
- FDNB26m
- Chemical Cleavage of Bonds29m
- Peptidases1h 6m
- Edman Degradation30m
- Edman Degradation Sequenator and Sequencing Data Analysis4m
- Edman Degradation Reaction Efficiency20m
- Ordering Cleaved Fragments21m
- Strategy for Ordering Cleaved Fragments58m
- Indirect Protein Sequencing Via Geneomic Analyses24m
- 6. Enzymes and Enzyme Kinetics13h 38m
- Enzymes24m
- Enzyme-Substrate Complex17m
- Lock and Key Vs. Induced Fit Models23m
- Optimal Enzyme Conditions9m
- Activation Energy24m
- Types of Enzymes41m
- Cofactor15m
- Catalysis19m
- Electrostatic and Metal Ion Catalysis11m
- Covalent Catalysis18m
- Reaction Rate10m
- Enzyme Kinetics24m
- Rate Constants and Rate Law35m
- Reaction Orders52m
- Rate Constant Units11m
- Initial Velocity31m
- Vmax Enzyme27m
- Km Enzyme42m
- Steady-State Conditions25m
- Michaelis-Menten Assumptions18m
- Michaelis-Menten Equation52m
- Lineweaver-Burk Plot43m
- Michaelis-Menten vs. Lineweaver-Burk Plots20m
- Shifting Lineweaver-Burk Plots37m
- Calculating Vmax40m
- Calculating Km31m
- Kcat46m
- Specificity Constant1h 1m
- 7. Enzyme Inhibition and Regulation 8h 42m
- Enzyme Inhibition13m
- Irreversible Inhibition12m
- Reversible Inhibition9m
- Inhibition Constant26m
- Degree of Inhibition15m
- Apparent Km and Vmax29m
- Inhibition Effects on Reaction Rate10m
- Competitive Inhibition52m
- Uncompetitive Inhibition33m
- Mixed Inhibition40m
- Noncompetitive Inhibition26m
- Recap of Reversible Inhibition37m
- Allosteric Regulation7m
- Allosteric Kinetics17m
- Allosteric Enzyme Conformations33m
- Allosteric Effectors18m
- Concerted (MWC) Model25m
- Sequential (KNF) Model20m
- Negative Feedback13m
- Positive Feedback15m
- Post Translational Modification14m
- Ubiquitination19m
- Phosphorylation16m
- Zymogens13m
- 8. Protein Function 9h 41m
- Introduction to Protein-Ligand Interactions15m
- Protein-Ligand Equilibrium Constants22m
- Protein-Ligand Fractional Saturation32m
- Myoglobin vs. Hemoglobin27m
- Heme Prosthetic Group31m
- Hemoglobin Cooperativity23m
- Hill Equation21m
- Hill Plot42m
- Hemoglobin Binding in Tissues & Lungs31m
- Hemoglobin Carbonation & Protonation19m
- Bohr Effect23m
- BPG Regulation of Hemoglobin24m
- Fetal Hemoglobin6m
- Sickle Cell Anemia24m
- Chymotrypsin18m
- Chymotrypsin's Catalytic Mechanism38m
- Glycogen Phosphorylase21m
- Liver vs Muscle Glycogen Phosphorylase21m
- Antibody35m
- ELISA15m
- Motor Proteins14m
- Skeletal Muscle Anatomy22m
- Skeletal Muscle Contraction45m
- 9. Carbohydrates7h 49m
- Carbohydrates19m
- Monosaccharides15m
- Stereochemistry of Monosaccharides33m
- Monosaccharide Configurations32m
- Cyclic Monosaccharides20m
- Hemiacetal vs. Hemiketal19m
- Anomer14m
- Mutarotation13m
- Pyranose Conformations23m
- Common Monosaccharides33m
- Derivatives of Monosaccharides21m
- Reducing Sugars21m
- Reducing Sugars Tests19m
- Glycosidic Bond48m
- Disaccharides40m
- Glycoconjugates12m
- Polysaccharide7m
- Cellulose7m
- Chitin8m
- Peptidoglycan12m
- Starch13m
- Glycogen14m
- Lectins16m
- 10. Lipids5h 49m
- Lipids15m
- Fatty Acids30m
- Fatty Acid Nomenclature11m
- Omega-3 Fatty Acids12m
- Triacylglycerols11m
- Glycerophospholipids24m
- Sphingolipids13m
- Sphingophospholipids8m
- Sphingoglycolipids12m
- Sphingolipid Recap22m
- Waxes5m
- Eicosanoids19m
- Isoprenoids9m
- Steroids14m
- Steroid Hormones11m
- Lipid Vitamins19m
- Comprehensive Final Lipid Map13m
- Biological Membranes16m
- Physical Properties of Biological Membranes18m
- Types of Membrane Proteins8m
- Integral Membrane Proteins16m
- Peripheral Membrane Proteins12m
- Lipid-Linked Membrane Proteins21m
- 11. Biological Membranes and Transport 6h 37m
- Biological Membrane Transport21m
- Passive vs. Active Transport18m
- Passive Membrane Transport21m
- Facilitated Diffusion8m
- Erythrocyte Facilitated Transporter Models30m
- Membrane Transport of Ions29m
- Primary Active Membrane Transport15m
- Sodium-Potassium Ion Pump20m
- SERCA: Calcium Ion Pump10m
- ABC Transporters12m
- Secondary Active Membrane Transport12m
- Glucose Active Symporter Model19m
- Endocytosis & Exocytosis18m
- Neurotransmitter Release23m
- Summary of Membrane Transport21m
- Thermodynamics of Membrane Diffusion: Uncharged Molecule51m
- Thermodynamics of Membrane Diffusion: Charged Ion1h 1m
- 12. Biosignaling9h 45m
- Introduction to Biosignaling44m
- G protein-Coupled Receptors32m
- Stimulatory Adenylate Cyclase GPCR Signaling42m
- cAMP & PKA28m
- Inhibitory Adenylate Cyclase GPCR Signaling29m
- Drugs & Toxins Affecting GPCR Signaling20m
- Recap of Adenylate Cyclase GPCR Signaling5m
- Phosphoinositide GPCR Signaling58m
- PSP Secondary Messengers & PKC27m
- Recap of Phosphoinositide Signaling7m
- Receptor Tyrosine Kinases26m
- Insulin28m
- Insulin Receptor23m
- Insulin Signaling on Glucose Metabolism57m
- Recap Of Insulin Signaling in Glucose Metabolism6m
- Insulin Signaling as a Growth Factor1h 1m
- Recap of Insulin Signaling As A Growth Factor9m
- Recap of Insulin Signaling1m
- Jak-Stat Signaling25m
- Lipid Hormone Signaling15m
- Summary of Biosignaling13m
- Signaling Defects & Cancer20m
- Review 1: Nucleic Acids, Lipids, & Membranes2h 47m
- Nucleic Acids 19m
- Nucleic Acids 211m
- Nucleic Acids 34m
- Nucleic Acids 44m
- DNA Sequencing 19m
- DNA Sequencing 211m
- Lipids 111m
- Lipids 24m
- Membrane Structure 110m
- Membrane Structure 29m
- Membrane Transport 18m
- Membrane Transport 24m
- Membrane Transport 36m
- Practice - Nucleic Acids 111m
- Practice - Nucleic Acids 23m
- Practice - Nucleic Acids 39m
- Lipids11m
- Practice - Membrane Structure 17m
- Practice - Membrane Structure 25m
- Practice - Membrane Transport 16m
- Practice - Membrane Transport 26m
- Review 2: Biosignaling, Glycolysis, Gluconeogenesis, & PP-Pathway3h 12m
- Biosignaling 19m
- Biosignaling 219m
- Biosignaling 311m
- Biosignaling 49m
- Glycolysis 17m
- Glycolysis 27m
- Glycolysis 38m
- Glycolysis 410m
- Fermentation6m
- Gluconeogenesis 18m
- Gluconeogenesis 27m
- Pentose Phosphate Pathway15m
- Practice - Biosignaling13m
- Practice - Bioenergetics 110m
- Practice - Bioenergetics 216m
- Practice - Glycolysis 111m
- Practice - Glycolysis 27m
- Practice - Gluconeogenesis5m
- Practice - Pentose Phosphate Path6m
- Review 3: Pyruvate & Fatty Acid Oxidation, Citric Acid Cycle, & Glycogen Metabolism2h 26m
- Pyruvate Oxidation9m
- Citric Acid Cycle 114m
- Citric Acid Cycle 27m
- Citric Acid Cycle 37m
- Citric Acid Cycle 411m
- Metabolic Regulation 18m
- Metabolic Regulation 213m
- Glycogen Metabolism 16m
- Glycogen Metabolism 28m
- Fatty Acid Oxidation 111m
- Fatty Acid Oxidation 28m
- Citric Acid Cycle Practice 17m
- Citric Acid Cycle Practice 26m
- Citric Acid Cycle Practice 32m
- Glucose and Glycogen Regulation Practice 14m
- Glucose and Glycogen Regulation Practice 26m
- Fatty Acid Oxidation Practice 14m
- Fatty Acid Oxidation Practice 27m
- Review 4: Amino Acid Oxidation, Oxidative Phosphorylation, & Photophosphorylation1h 48m
- Amino Acid Oxidation 15m
- Amino Acid Oxidation 211m
- Oxidative Phosphorylation 18m
- Oxidative Phosphorylation 210m
- Oxidative Phosphorylation 310m
- Oxidative Phosphorylation 47m
- Photophosphorylation 15m
- Photophosphorylation 29m
- Photophosphorylation 310m
- Practice: Amino Acid Oxidation 12m
- Practice: Amino Acid Oxidation 22m
- Practice: Oxidative Phosphorylation 15m
- Practice: Oxidative Phosphorylation 24m
- Practice: Oxidative Phosphorylation 35m
- Practice: Photophosphorylation 15m
- Practice: Photophosphorylation 21m
Isoelectric Point of a Peptide: Study with Video Lessons, Practice Problems & Examples
 Created using AI
Created using AITo calculate the isoelectric point (pI) of a peptide, average the two pKa values that sandwich the pH where the predominant structure has a neutral net charge. Identify the seven amino acids with ionizable R groups using the mnemonic "yucky crazy dragons knights riding horses." Order the pKa values from smallest to largest, determine the net charge at various pH levels, and find the pH range with a net charge of zero. The pI is calculated as
Isoelectric Point of a Peptide
Video transcript
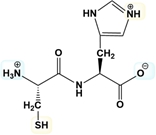
Estimate the isoelectric point of the following dipeptide in the figure.
Calculate the approximate pI of the peptide: C-G-E-K.
Draw in the R-groups for the following peptide & calculate the pI: ATLDAG.
Here’s what students ask on this topic:
What is the isoelectric point (pI) of a peptide?
The isoelectric point (pI) of a peptide is the pH at which the peptide has a net charge of zero. To calculate the pI, you need to average the two pKa values that sandwich the pH where the predominant structure of the peptide is neutral. This involves identifying the ionizable groups in the peptide, ordering their pKa values from smallest to largest, and determining the net charge at various pH levels. The pI is then calculated using the formula:
 Created using AI
Created using AIHow do you calculate the isoelectric point of a peptide?
To calculate the isoelectric point (pI) of a peptide, follow these steps:
- Identify the ionizable groups in the peptide and their pKa values.
- Order the pKa values from smallest to largest.
- Determine the net charge of the peptide at various pH levels between each pair of adjacent pKa values.
- Find the pH range where the peptide has a net charge of zero.
- Average the two pKa values that sandwich this pH range using the formula:
 Created using AI
Created using AIWhy is the isoelectric point important in biochemistry?
The isoelectric point (pI) is crucial in biochemistry because it affects the solubility and behavior of peptides and proteins in different pH environments. At the pI, a peptide or protein has no net charge, which can lead to precipitation or reduced solubility. Understanding the pI helps in protein purification, crystallization, and understanding protein interactions. It also plays a role in techniques like isoelectric focusing, where proteins are separated based on their pI values.
 Created using AI
Created using AIWhat are the seven amino acids with ionizable R groups?
The seven amino acids with ionizable R groups are:
- Tyrosine (Y)
- Cysteine (C)
- Aspartic acid (D)
- Glutamic acid (E)
- Lysine (K)
- Arginine (R)
- Histidine (H)
These amino acids can donate or accept protons, affecting the net charge of the peptide at different pH levels. A mnemonic to remember them is "yucky crazy dragons knights riding horses."
 Created using AI
Created using AIHow does the microenvironment affect the pKa values of amino acid residues in a peptide?
The microenvironment within a peptide can significantly affect the pKa values of amino acid residues. Factors such as the local polarity, hydrogen bonding, and the presence of nearby charged groups can shift the pKa values from those of free amino acids. This is why the pI and net charge of a peptide can only be estimated and are best determined experimentally. When calculating the pI, it's essential to use the correct set of pKa values specific to amino acid residues within peptides.
 Created using AI
Created using AI